Abstract
Colorectal cancer is the third most commonly occurring cancer in men and the second in women. The global burden of colorectal cancer is projected to increase to over 2 million new cases with over 1 million deaths within the next 10 years, and there is a great need for new compounds with novel mechanisms of action. Our group has developed protein kinase C (PKC)–modulating isophthalic acid derivatives that induce cytotoxicity toward human cervical and prostate cancer cell lines. In this study, we investigated the effects of 5-(hydroxymethyl)isophthalate 1a3 (HMI-1a3) on colorectal cancer cell lines (Caco-2, Colo205, and HT29). HMI-1a3 inhibited cell proliferation, decreased cell viability, and induced an apoptotic response in all studied cell lines. These effects, however, were independent of PKC. Using serine/threonine kinome profiling and pharmacological kinase inhibitors, we identified activation of the cAMP/PKA pathway as a new mechanism of action for HMI-1a3–induced anticancer activity in colorectal cancer cell lines. Our current results strengthen the hypothesis for HMI-1a3 as a potential anticancer agent against various malignancies.
SIGNIFICANCE STATEMENT Colorectal cancer (CRC) is a common solid organ malignancy. This study demonstrates that the protein kinase C (PKC)–C1 domain–targeted isophthalatic acid derivative 5-(hydroxymethyl)isophthalate 1a3 (HMI-1a3) has anticancer activity on CRC cell lines independently of PKC. We identified PKA activation as a mechanism of HMI-1a3–induced anticancer effects. The results reveal a new anticancer mechanism of action for the partial PKC agonist HMI-1a3 and thus provide new insights for the development of PKC and PKA modulators for cancer therapy.
Introduction
Worldwide, approximately 10% of all diagnosed cancers and cancer-related deaths are because of colorectal cancer (CRC) (Bray et al., 2018). Although the prognosis of patients with CRC has improved during the past decades, the 5-year survival rate remains around 50%–65% (Brenner et al., 2012; Bray and Soerjomataram, 2015). Therefore, new treatment regimens to combat advanced disease with alternative therapeutic strategies are needed.
Protein kinases play a key role in tumor growth and progression; thus, they have been an attractive target for drug discovery for decades. As of August 20, 2021, the US Food and Drug Administration has approved 73 small molecules targeting protein kinases (Cohen et al., 2021; FDA, 2021). These compounds are mainly protein kinase inhibitors, since protein kinase cascades are frequently hyperactivated in cancer.
A majority of the drugs either directly or allosterically target the ATP-binding site of a protein kinase (Roskoski, 2015). Since the ATP-binding site among various protein kinases shares structural similarity, most if not all protein kinase inhibitors inhibit several kinases at the same time. Even so, usually the therapeutic effect is remarkable. Targeting regulatory domain of intracellular protein kinases has been less studied, but we and others have demonstrated protein kinase C (PKC)–C1 domain as a potential target (Bessa et al., 2018; Jäntti et al., 2018). Protein kinase A (PKA) and PKC are serine/threonine kinases involved in CRC development (Spindler et al., 2009; Kisslov et al., 2012). Several lines of evidence suggest that activation of PKA inhibits progression of CRC (Zhao et al., 2021). Investigational inhibitors of PKC targeting the ATP-binding site have failed in clinical trials for CRC (Ouaret and Larsen, 2014). In the case of PKC, rather than inhibiting its activity, therapeutic strategies should aim to restore its activity (Antal et al., 2015). Among more than 500 PKC mutations in biopsies from patients with cancer, detailed characterization of 46 representative mutations showed that cancer-associated point mutations in PKC were loss-of-function mutations. Similar results have also been reported by other research groups (Craven and DeRubertis, 1994; Pongracz et al., 1995; Suga et al., 1998; Dowling et al., 2016). These findings substantially change the generally believed role of PKC in tumor promotion and progression. Considering this, it seems reasonable that instead of inhibiting PKC, we should aim at preserving or increasing its activity to treat cancer.
Our group has previously developed isophthalic acid derivatives binding to the C1-domain of classic and novel PKCs, modulating their activity without inducing downregulation of PKC (Boije af Gennäs et al., 2009; Jäntti et al., 2018). They have been demonstrated to possess PKC-mediated neuroprotective effects against neuroinflammation (Sarajärvi et al., 2018) and antifibrotic effects on cardiac fibroblasts (Karhu et al., 2021). They also inhibit the proliferation of HeLa human cervical cancer cells and induce senescence or apoptosis in PC3, DU145, and LNCaP prostate cancer cell lines (Talman et al., 2011; Jäntti et al., 2018). Since there is strong evidence suggesting that PKCs play a tumor-suppressing role especially in colorectal cancer (Choi et al., 1990; Antal et al., 2015; Dowling et al., 2016), yet the anticancer effects observed in the previous studies have not been fully PKC-mediated, we aimed to unravel the effects and mechanisms of 5-(hydroxymethyl)isophthalate 1a3 (HMI-1a3) in colorectal cancer cell lines Caco-2, Colo205, and HT-29.
Materials and Methods
Compounds and Reagents
HMI-1a3 was synthesized at the Division of Pharmaceutical Chemistry and Technology, Faculty of Pharmacy, University of Helsinki, as described previously (Boije af Gennäs et al., 2009). High-definition NMR spectrometery (Bruker Ascend 400 MHz-Avance III; Bruker Corporation, Billerica, MA) was used to verify the identity of the compounds. Exact mass and purity (>95%) of each batch was confirmed by liquid chromatography–mass spectrometry with a Waters Acquity UPLC system (Waters, Milford, MA) that was equipped with an Acquity UPLC BEH C18 column (1.7 µm, 50 × 2.1 mm; Waters, Dublin, Ireland) and Acquity PDA detector. Waters Synapt G2 HDMS mass spectrometer (Waters, Milford, MA) was used with an electrospray ion source in positive mode. The PKC activator phorbol 12-myristate 13-acetate (PMA), the pan-PKC inhibitor Gö6983, the conventional-PKC inhibitor Gö6976, and the glycogen synthase kinase 3 (GSK-3) inhibitor CHIR99021 were from Sigma-Aldrich (Steinheim, Germany). Ca2+/calmodulin-dependent protein kinase type 2 (CaMK 2) inhibitor KN93, c-Jun N-terminal kinase (JNK) inhibitor SP600125, mitogen-activated protein kinase kinase 1/2 inhibitor U0126, mitogen-activated protein kinase-activated protein kinase 2/p38-activated protein kinase (PRAK) inhibitor PF3644022, p38α and p38β [mitogen-activated protein kinase (MAPK)] inhibitor SB203580, phospholipase C inhibitor U73122, PKA inhibitor KT5720, PKA inhibitor cAMPS-Rp, PKG inhibitor KT5823, and staurosporine were from Tocris Bioscience (Bristol, UK).
Trans-Blot Turbo Midi PVDF Transfer Packs and 12% Mini-Protean TGX Stain-Free Protein Gels used for Western blotting were purchased from Bio-Rad Laboratories (Hercules, CA). The Pierce BCA Protein Assay Kit and SuperSignal West Femto Maximum Sensitivity Substrate reagents were from Thermo Fisher Scientific (Paisley, UK). Primary antibodies were acquired from the following sources: E2F1 (#3742, 1:2000; Cell Signaling Technology, Danvers, MA), p21 Waf1/Cip1 (#2946, 1:1000; Cell Signaling Technology), phospho–vasodilator-stimulated phosphoprotein (VASP) (Ser 157) (# 365564, 1:200; Santa Cruz Biotechnology, Dallas, TX), phospho- VASP (Ser 239) (# 3114, 1:1000; Cell Signaling Technology), cleaved poly(ADP-ribose) polymerase (PARP) (#9541, 1:1000; Cell Signaling Technology), VASP (#46668, 1:1000; Santa Cruz Biotechnology), anti–β-actin (#4967, 1:1000; Cell Signaling Technology), and Anti–Glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH) Antibody, clone 6C5 (MAB374, 1:10000; Sigma-Aldrich). The secondary horseradish peroxidase–linked antibodies: goat anti-rabbit IgG (#7074, 1:2000) and horse anti-mouse IgG (#7076, 1:2000) were bought from Cell Signaling Technology.
For immunofluorescence staining and high-content analysis (HCA), 5-bromo-2′-deoxyuridine (BrdU) was acquired from Abcam (Cambridge, UK), monoclonal rat anti-BrdU (#ab6326) was acquired from Abcam, and the Alexa Fluor 647–conjugated goat anti-rat lgG was acquired from Thermo Fisher Scientific (#A21247). The DNA stain 4′,6-diamidino-2-phenylindole (DAPI) was from Sigma-Aldrich. CellEvent Caspase-3/7 Green Detection Reagent was purchased from Invitrogen (Carlsbad, CA).
Cell Culture
The human colorectal cancer cell lines Caco-2 (colorectal adenocarcinoma; male), Colo205 (colorectal adenocarcinoma; male), and HT-29 (colorectal adenocarcinoma; female) were purchased from the American Type Culture Collection (Manassas, VA). Colo205 cells were cultured in RPMI1640 medium (#1060120; MP Biomedicals, Santa Ana, CA), and Caco-2 and HT-29 cells were cultured in Dulbecco’s modified Eagle’s medium (D-7777; Sigma-Aldrich). For all cell lines, the basal media were supplemented with 10% FBS, 100 units/ml penicillin, and 100 µg/ml streptomycin (all sourced from Gibco). Cell cultures were maintained at 37°C in a humidified atmosphere of 5% CO2. For viability and HCA, cells were plated on Corning Costar 96-well plates, and for immunoblotting and Ser/Thr kinase profiling, cells were plated on plastic six-well plates.
Cell Viability Assays
Cell viability was determined by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) reduction assay, and cell membrane integrity was determined using lactate dehydrogenase (LDH) release assay from cell culture media as previously described (Tarvainen et al., 2019). Briefly, the cells seeded onto 96-well plates (10,000 cells/well in serum-supplemented media) were incubated with the compounds in normal cell culture medium for 24 hours. The LDH assay was carried out with 50-µl samples of culture medium from each well. Fifty microliters of substrate solution was added, and after a 30-minute incubation at room temperature (RT) in the dark, the reaction was stopped by adding 50 µl of 1 M acetic acid. Absorbance was measured at 490 nm using Victor2 plate reader (PerkinElmer, Turku, Finland). Spontaneous, maximal, and background LDH activity was quantified from cell culture medium of untreated cells, medium of cells lysed with 0.9% Triton X-100, and medium without cells, respectively. For quantification of metabolic activity, MTT solution was added to the cells at a final concentration of 0.5 mg/ml, and the cells were grown for an additional 2 hours in cell culture conditions. The cell culture media were then aspirated, and DMSO (200 μl) was added to dissolve the formazan crystals. Absorbance was measured at 550 nm, subtracting absorbance at 650 nm as background. Experiments were performed in biologic triplicates.
Immunoblotting
For the analysis of protein expression or phosphorylation, cells were plated at 4.0 × 105 cells/well on six-well plates and left to attach for 24 hours. The cells were then exposed to the compounds (to study cleaved PARP, E2F1, and p21 Waf1/Cip1), or the medium was changed to a serum-free cell culture medium for 24 hours prior to the compound exposure (to study VASP). The cells were treated with the test compounds for the indicated times, after which the medium was removed, and the cells were washed twice with ice-cold PBS. Cells were then harvested in ice-cold lysis buffer containing 1 mM EDTA, 150 mM NaCl, 0.25% NP-40, 1% Triton X-100, 10 mM Tris-HCl (pH 6.8) and supplemented with complete protease inhibitors (Roche, Mannheim, Germany) and PhosSTOP phosphatase inhibitors (Roche). Cell homogenates were centrifuged at 13,000 ×g for 4 minutes at 4°C, and the supernatants were collected for further analysis. Protein concentrations were determined with bicinchoninic acid protein assay kit. Equal amounts of protein (10 µg/lane) were resolved on a 12% Mini-Protean TGX Stain-Free Protein Gel in reducing conditions and transferred to poly(vinylidene difluoride) membrane by the Trans-Blot Turbo transfer system. To prevent nonspecific reactions from occurring, membranes were then blocked with 5% dry milk in 0.1% Tween 20 in Tris-buffered saline for 1 hour at RT, after which they were incubated overnight at 4°C in a shaker with primary antibodies in blocking buffer overnight. On the next day, the membranes were washed with 0.1% Tween 20 in Tris-buffered saline and incubated with blocking buffer containing horseradish peroxidase–linked secondary antibody for 2 hours at RT. Detection of secondary antibodies was done using a chemiluminescent substrate utilizing ChemiDoc XRS+ imaging system (Bio-Rad Laboratories). For quantification, the optical densities of the immunoreactive protein bands were analyzed using the ImageJ software (https://imagej.net/Downloads). The optical densities were first normalized to the corresponding loading control (GAPDH or β-actin) and then to the concurrent control on the same membrane. The experiments were replicated three times with two wells per condition in each experiment.
Serine/Threonine Kinome Profiling
The Ser/Thr kinase (STK) profile of HMI-1a3 in Colo205 was determined by PamStation12 and STK PamChips (PamGene, 's-Hertogenbosch, The Netherlands). The fluorescent platform measures phosphorylation of specific substrate peptides embedded on multiplex chip arrays by active kinases in the sample in the presence of ATP. Each array displays 144 immobilized phosphorylatable peptides of 15 amino acids, which correspond to known or reputed phosphorylation sites of human proteins derived from the literature and correlated with one or several upstream kinases.
The STK activity profiling was conducted according to manufacturer’s standard protocol using reagents from Pamgene. The cells were grown on six-well plates and exposed to the compounds for 30 minutes or 2 hours. After removal of the medium from the plate, the cells were washed with ice-cold PBS and lysed on ice with M-PER Mammalian Protein Extraction Reagent supplemented with Halt Phosphatase and Halt Protease inhibitor cocktails (all from Thermo Fisher Scientific). The lysate was centrifuged at 16,000 ×g for 15 minutes at 4°C, and the supernatant was immediately flash-frozen in liquid nitrogen and then stored at −80°C. Total protein levels were determined by Bradford assay, and the STK Basic Mix was composed of 1 µg of the lysate, 4 µl of 10 × protein reaction buffer stock, 0.4 µl of 100 × bovine serum albumin stock, 4.0 µl of 4 mM ATP, and 0.46 µl STK antibody mix. Total volume of the STK Basic Mix was adjusted to 40 µl by adding distilled water. The detection mix consisted of 3 µl of 10× antibody buffer, 0.4 µl fluorescein isothiocyanate–labeled STK antibody, and 26.6 µl distilled water. The fluorescence was detected using a CCD camera, and signals were integrated into one single value for each peptide with software-based image analysis. Raw image analysis was conducted using Evolve2 (Pamgene) software, with comparative analysis done in BioNavigator Analysis software (Pamgene). The mean phosphorylation intensities (after background correction) of each phospho-Ser/Thr peptide were normalized to the corresponding mean signal intensity of DMSO control treatment and expressed as log2 fold-change ratio. Experiments were performed in biologic triplicates.
High-Content Analysis
For HCA of proliferation and caspase activation, the staining, imaging, and analyzing of cells were done as described previously (Karhu et al., 2018). The cells were treated with the test compounds for 24 hours, and 10 µM BrdU was added to the culture medium for the last 1 hour prior to fixation with 4% paraformaldehyde for 15 minutes at RT. Immunofluorescence staining was done at RT as follows. The cells were permeabilized with 0.1% Triton X-100 for 10 minutes. DNA hydrolysis was done with 2 M hydrochloric acid for 30 minutes, and subsequent neutralization was done with 0.1 M sodium borate (pH 8.5) for 30 minutes. After a 45-minute blocking with 4% FBS in PBS, the cells were incubated with anti-BrdU antibody (1:250) for 1 hour, which was followed by three 5-minute washes with PBS and a 45-minute incubation with Alexa Fluor 647–conjugated secondary antibody (1:200) and DAPI (5 µg/ml) for 45 minutes. For the analysis of caspase activation, the cells were treated with the compounds for 4 hours and then incubated with 5 µM CellEvent Caspase-3/7 Green Detection Reagent solution in PBS with 5% FBS for 50 minutes before fixation.
The samples were imaged with ImageXpress Nano high-content imaging system (Molecular Devices, Winnersh, UK). Sixteen images per well were collected using a 10× objective to analyze >100 cells from each well. The images were analyzed with the MetaXpress software, as follows: The nuclei were first identified based on DNA staining (DAPI), the nuclear fluorescence intensity at 647 nm was quantified as a measure of BrdU, and the nuclear fluorescence intensity at 488 nm was quantified as a measure of caspase 3/7 activity.
Statistics
Statistical analyses and IC50 value calculations were performed using GraphPad Prism (version 7.04) software for Windows (GraphPad Software, La Jolla, CA, www.graphpad.com). Control refers to incubation with vehicle (0.1% DMSO). N refers to the number of independent experiments, for which two or more parallel samples (technical replicates) were averaged to produce N = 1. Data are presented as the mean of independent experiments, the number of which is indicated in the corresponding figure legends.
Results
HMI-1a3 Decreases the Viability of Colorectal Cancer Cell Lines Concentration-Dependently but in a PKC-Independent Manner
Caco-2, Colo205, and HT-29 cells were treated with HMI-1a3 at concentrations ranging from 0.1 to 20 μM for 24 hours, and cell viability was analyzed with the MTT assay (Fig. 1A). HMI-1a3 decreased the viability of all the studied cell lines in a concentration-dependent manner after 24-hour exposure. IC50 values were 2.9, 0.69, and 0.64 µM for Caco-2, Colo205, and HT29 cell lines, respectively. HMI-1a3 did not induce cell membrane damage as determined by the LDH activity in cell culture medium (Supplemental Tables 1, 2, and 3). Simultaneous treatment with the pan-PKC inhibitor Gö6983 (1 µM) did not attenuate the HMI-1a3–induced CRC cell viability reduction (Fig. 1B).
Effects of HMI-1a3 on CRC cell line viability. (A) HMI-1a3 decreased the viability of Caco-2, Colo205, and HT29 CRC cells concentration-dependently. (B) The effect was not reversed by PKC inhibitor Gö6983 (1 µM). Cell viability was measured in the MTT assay after 24-hour exposure with HMI-1a3. The data are presented as mean (A) ± S.E.M. and (B) + S.E.M. N = 3, *P < 0.05, and **P < 0.01 vs. control (CTRL), using ANOVA followed by Dunnett’s test.
HMI-1a3 Induces Antiproliferative and Proapoptotic Effects in Colorectal Cancer Cell Lines
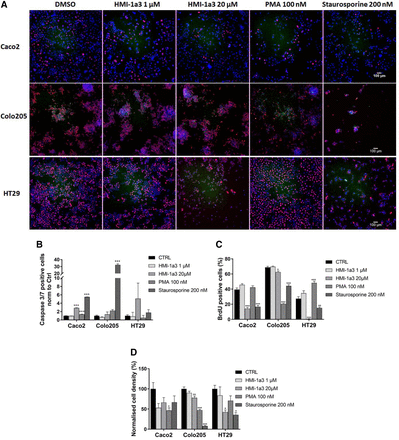
HCA and PARP-immunoblotting were used to study whether the decreased cell viability was because of the effects of HMI-1a3 on the proliferation and apoptosis of the CRC cell lines (Fig. 2; Supplemental Fig. 1). The potent PKC activator PMA was used to assess the possible role of PKC and staurosporine, a widely used inducer of apoptosis, as a positive control. On Caco-2 cells, HMI-1a3 did not have any clear effect on caspase 3/7 activity on after 24-hour exposure, whereas on Colo205 and HT29 cells, 20 µM HMI-1a3 increased the proportion of caspase-positive cells (Fig. 2B). HMI-1a3 at 20 µM decreased the percentage of BrdU-positive cells significantly in all cell lines studied (Fig. 2C). Both PMA and staurosporine had inconsistent effects in different cell lines (Fig. 2).
Effects of HMI-1a3 on cell proliferation and caspase activation in Caco-2, Colo205, and HT29 CRC cell lines. After a 24-hour compound incubation, the cells were incubated with BrdU (1 hour) and detection reagent for activated caspase 3/7 (50 minutes) (green) and subsequently fixed and stained for DNA (DAPI) (blue) and BrdU (magenta). (A) Representative images from each cell line and treatment are shown. Individual color channels were adjusted to enhance brightness and contrast for all representative images. Normalized number of cells positive for fluorescent caspase 3/7 activity reporter (B), quantification of the proportion of cells positive for BrdU (C), and normalized number of cells (D) are presented as mean + S.E.M. N = 3, *P < 0.05, and **P < 0.01 vs. control (CTRL), using ANOVA followed by Dunnett’s test.
Since the number of caspase 3/7–positive cells was very low, we hypothesized that the treatments or the fixing procedure may have detached dead cells and thus reduced the number of cells in the HCA results (Fig. 2, B and D). To abolish the uncertainty, we decided to verify the apoptosis by detecting the PARP cleavage with immunoblotting where a clear increase in the PARP cleavage in all the cell lines tested was observed (Supplemental Fig. 1). The CRC cells did not show senescent-like morphology or altered E2F1 or p21 expression (Supplemental Fig. 2), which ruled out the senescence as a cause of reduced viability in response to HMI-1a3, as previously observed in DU145 and PC3 prostate cancer cells (Jäntti et al., 2018).
Early Effects of HMI-1a3 on Kinase Activity of Colo205 Cells
Since the HMI-1a3–induced decline in CRC cell viability was PKC-independent, we sought to profile the effect of HMI-1a3 on the activity of STK on Colo205 cells after 30-minute and 2-hour exposures using PamStation 12. We hypothesized that the top-ranked kinases would represent the most relevant non-PKC mediators of HMI-1a3 anticancer activity. Kinase profiling revealed increased activation of several kinases, whereas no decrease of kinase activity was observed (Supplemental Figs. 3 and 4). Kinases were ranked based on median final score, and a higher specificity score indicates a higher likelihood of the corresponding kinase contributing to the observed phosphorylation changes (Tables 1 and 2). The changes in the kinase activity, however, were quite modest in both time points compared with control.
Ten top-ranked kinases after 30-minute exposure to 20 µM of HMI-1a3 compared with control according to the upstream kinase analysis
The obtained kinase profile was reflected on literature to delineate the potential pathways of the observed effects of HMI-1a3 on cancer cell viability. Extracellular signal-regulated kinase 7, which was activated after a 30-minute HMI-1a3 treatment, has been shown to serve as both a proto-oncogene and a tumor suppressor (Lau and Xu, 2018). CaMK 2 (Chen et al., 2017) and cyclin-dependent kinase 14 (Zhou et al., 2014) have been shown to be overexpressed in human colon cancer and contribute to invasiveness. The most upregulated kinases were different after 2 hours compared with 30 minutes (Table 2; Supplemental Fig. 4), and there was no clear correlation or trend between the time points. One of the most activated kinases after the 2-hour time point was cyclin-dependent kinaselike (CDKL) 2. Downregulation or loss of CDKL2 has been associated with several cancer cell types, including colorectal cancer, and its overexpression suppresses cells growth and invasion (Fang et al., 2018). This information suggests that CDKL2 activation by HMI-1a3 could mediate the observed decrease in viability in colorectal cancer cells. The other three most activated kinases, intestinal cell kinase (Whitworth et al., 2012), α-protein kinase (Chen et al., 2019), and CDKL5 (Jiang et al., 2019), are associated with tumor growth and aggressiveness. Being among the most activated kinases, according to the upstream kinase interpretation, cyclic AMP–dependent PKA and cyclic GMP–dependent PKG also demonstrated high specificity score or consistency in activity on both time points (Tables 1 and 2; Supplemental Figs. 3 and 4).
Ten top-ranked kinases after 2-hour exposure to 20 µM of HMI-1a3 compared with control according to the upstream kinase analysis
Unraveling the Mediators of HMI-1a3–Induced Effects in Colo205 Cells
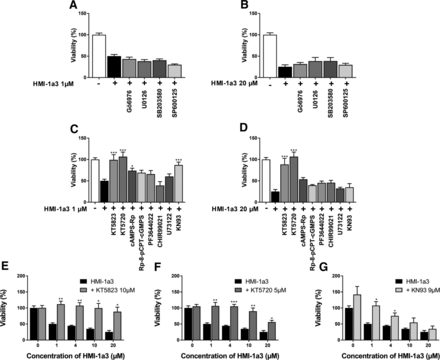
HMI-1a3 has been designed to bind PKC, and in addition to PKCα and –δ (Boije af Gennäs et al., 2009), it has been demonstrated to bind to β2-chimaerin, PKD1, and myotonic dystrophy kinase–related CDC42-binding kinase-α (Talman et al., 2014), all of which also contain the C1 structure. However, the effect of HMI-1a3 on CRC viability was not antagonized by the pan-PKC inhibitor Gö6983 (1 µM), the conventional PKC and PKD1 inhibitor Gö6976 (2 µM), or the inhibitors of main MAPK families MEK1/2, JNK, and p38α/p38β (10 µM U0126, 10 µM SP600125, and 5 µM SB20358, respectively), which are well known downstream mediators of PKC activation (Fig. 3, A and B). Therefore, we next tested a set of kinase inhibitors based on the STK profiling to find the possible mediators of the observed HMI-1a3–induced effects.
Effect of different inhibitors on the HMI-1a3 compromised viability of Colo205 colorectal cancer cells. (A and B) PKC inhibitors Gö6983 and Gö6976, MEK1/2 inhibitor U0126, MAPK inhibitor SB203580, or JNK inhibitor SP600125 did not antagonize the effect of 1 (A) or 20 µM (B) HMI-1a3. PKG KT5823 (C–E), PKA inhibitors KT5720 and cAMPS-Rp (C, D, and F), and CaMK 2 inhibitor KN93 (C, D, and G) inhibited the effects of HMI-1a3 fully or partially, whereas the PKG inhibitor Rp-8-pCPT-cGMPS, mitogen-activated protein kinase-activated protein kinase 2/PRAK inhibitor PF3644022, GSK-3 inhibitor CHIR99021, or phospholipase C inhibitor U73122 did not (A and B). Cell viability was measured utilizing MTT assay. Data are presented as mean + S.E.M. For A and B, N = 3, *P < 0.05, **P < 0.01, and ***P < 0.001 vs. HMI-1a3 treatment, using ANOVA followed by Dunnett’s test. For C–G, N = 3, *P < 0.05, **P < 0.01, and ***P < 0.001 vs. the corresponding HMI-1a3 treatment, using Student’s t test.
The PKG, PKC, and PKA inhibitor (Inhibition constant values 0.23, 4, and >10 μM, respectively) KT5823 reversed the HMI-1a3–induced decrease in cell viability at 10 µM concentration (Fig. 3, C–E). The more-specific inhibitors of PKA KT5720 (5 µM) and cAMPS-Rp (100 µM) attenuated the effects of HMI-1a3 to some extent yet not as strongly as KT5823, whereas the PKG inhibitor (Rp-8-pCPT-cGMPS, 250 µM) did not antagonize the observed effects of HMI-1a3 (Fig. 3, C and D). Also, the CaMK 2 inhibitor KN93 (9 µM) seemed to block the viability decreasing effect of 1 µM HMI-1a3 but not that of 20 µM HMI-1a3 (Fig. 3, C and D). Phospholipase C inhibitor U73122 (5 µM) did not mitigate the effect of HMI-1a3. Based on these findings, KT5823, KT5720, and KN93 were chosen for a more profound concentration-response experiment, wherein it was shown that KT5823 and KT5720 inhibited the effect of HMI-1a3 on cell viability significantly, whereas KN93 seemed to increase the metabolic activity itself, which made it difficult to distinguish its possible HMI-1a3–inhibiting effect (Fig. 3, E–G).
Bain et al. (2003) showed in their kinase-inhibitor screening that instead of PKG, PKC, or PKA, KT5823 inhibits PRAK and GSK-3β. They also suggested that there might be significant interbatch differences explaining the lack of PKG-inhibiting properties. Because of this, we studied the effects of PRAK and GSK-3 inhibitors (250 µM PF3644022 and 12 µM CHIR99021 respectively), but they did not affect the decrease in cell viability (Fig. 3, C and D). Furthermore, we did not observe any interbatch differences between compound batches used in our assays.
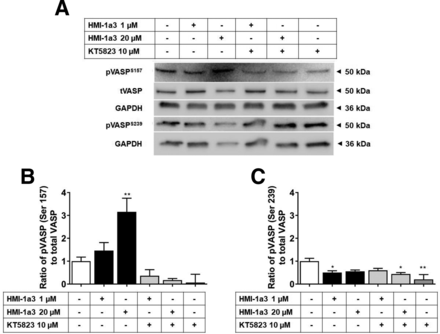
To characterize the possible role of PKA and PKG, the phosphorylation of VASP after 30 minutes was investigated using immunoblotting. Since several phosphorylation sites of VASP have been identified, we studied both the major PKA phosphorylation site Ser157 and the major PKG phosphorylation site Ser239 (Smolenski et al., 1998). A 30-minute exposure to 1 and 20 µM HMI-1a3 increased the phosphorylation of the VASP Ser157, which indicated that HMI-1a3 treatment indeed activates PKA in Colo205 cells (Fig. 4). An opposite effect was observed on the phosphorylation of VASP at Ser239. KT5823 inhibited HMI-1a3–induced VASP phosphorylation at Ser157.
HMI-1a3 increases the phosphorylation of VASP at serine 157 but not at serine 239 in Colo205 CRC cell line after 30-minute treatment. (A) Representative Western blots and quantifications of the proportion of (B) phosphorylated VASP (Ser 157) and (C) VASP (Ser 239) in response to HMI-1a3 and KT5823. Data are presented as mean + S.E.M. N = 3, *P < 0.05, and **P < 0.01 vs. control, using ANOVA followed by Dunnet’s test. pVASP, phosphorylated VASP; tVASP, total VASP.
Discussion
Even though PKC has been generally considered as an oncogenic kinase, several studies have demonstrated that loss of PKC activity is often associated with cancer. In these cases, PKC isoforms function as tumor suppressors rather than promoters (Newton, 2018). Consistent with these findings, the development of PKC activators not inducing downregulation of PKC protein levels should be the basis for a potential new mechanism of action for an anticancer drug (Oliva et al., 2008; Boije af Gennäs et al., 2009; Bessa et al., 2018).
In our previous studies, we showed that the derivative of isophthalic acid HMI-1a3 binds to the C1 domain and activates PKCs without inducing downregulation (Boije af Gennäs et al., 2009). HMI-1a3 has been shown to inhibit the proliferation of HeLa cervical cancer cells (Talman et al., 2011) and DU145, PC3, and LNCaP prostate cancer cells (Jäntti et al., 2018). HMI-1a3 induced apoptosis in LNCaP cells and senescence in DU145 and PC3 cells. In HeLa, PC3, and DU145 cells, the pan-PKC inhibitor Gö6983 did not attenuate the effects of HMI-1a3, whereas in LNCaP cells, its effect seems to be mediated by PKC (Talman et al., 2011, 2014; Jäntti et al., 2018).
In the current study, we characterized the effects of HMI-1a3 on colorectal cancer cells in vitro, since it has been previously suggested that PKC acts as a tumor suppressor in colorectal cancer (Doi et al., 1994; Goldstein et al., 1995; Lee et al., 2010; Dowling et al., 2016). HMI-1a3 decreased the viability and induced apoptosis in all tested CRC cell lines; however, the observed effects were not attenuated with the PKC inhibitors Gö6983 or Gö6976. Since it seemed that the effects of isophthalic acid derivative HMI-1a3 were not mediated by PKC or the downstream mediators of PKC signaling, we investigated its mechanism of action utilizing a Pamgene serine/threonine kinome–profiling assay. It is important to acknowledge that the kinome profile relies on literature and databases and it is an interpretation from the phosphopeptide read-out. The profile of the early effects of HMI-1a3 did not include any of the known C1 domain–containing proteins (Kazanietz, 2002; Griner and Kazanietz, 2007; Talman et al., 2014), nor were there any clear trends between the two time points. The results, however, suggest that PKA and PKG play a role, even though cAMP/PKA-related or cGMP/PKG-related kinases were not among the top hit kinases in the latter time point. We subsequently showed that a nonspecific PKG inhibitor KT5823 and two different PKA inhibitors attenuated the viability decreasing effects of HMI-1a3. More detailed investigation revealed that HMI-1a3 increased the phosphorylation of VASP at Ser157, whereas it surprisingly decreased the phosphorylation at Ser239. Ser157 is a preferred phosphorylation site for PKA, and Ser239 is the preferential phosphorylation site for PKG; thus, this finding supports a preferential involvement of cAMP-PKA pathway in the effects of HMI-1a3. In some cases, VASP Ser157 is also phosphorylated by PKC (Chitaley et al., 2004) and PKD1 and 2 (Doppler et al., 2013, 2015), but here the effect was shown to be PKA-mediated.
Activation of cAMP-PKA signaling and phosphorylation of VASP Ser157 has tumor-suppressive and tumor-promoting effects depending on the tumor types and tissue of origin (Zhang et al., 2020; Ali et al., 2021). Activation of the cAMP-PKA signaling with the cAMP analog 8-Cl-cAMP has been demonstrated to inhibit the growth of CRC, breast cancer, lung cancer, and leukemia in vitro and in vivo (Zhang et al., 2020). These results are consistent with our current finding that the anticancer activity of HMI-1a3 was inhibited by the cAMP analog cAMPS-Rp, which inhibits PKA. The significance of VASP Ser157 phosphorylation in the regulation of cancer cell signaling is controversial. The anticancer drug imatinib restores the phosphorylation of VASP Ser157 and decreases VASP and breakpoint cluster region–abelson protein interaction in imatinib-responsive, chronic myelogenous leukemia (Bernusso et al., 2015). In prostate cancer cells, Ser157 phosphorylation of VASP correlates with cell motility and has been suggested to be a marker for metastatic progression potential (Hasegawa et al., 2008). In a recent study, VASP Ser157 phosphorylation was associated with enhanced colon cancer cell clonogenicity and migration in vitro and growth into tumors in nude mice (Ali et al., 2021). However, phosphorylation of VASP Ser239 phosphorylation had an opposite effect. Our results with cAMP analogs and HMI-1a3 support a hypothesis that there is a certain balance in the phosphorylation of VASP Ser157 and VASP Ser239, which contributes to the tumor-suppressing effect of PKA activation. This may explain the differential findings of tumor cell line–suppressive effects of PKA signaling in some but not all experimental settings.
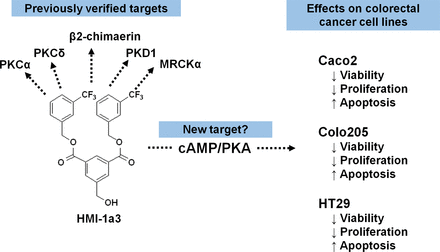
Previously established antiproliferative and proapoptotic effects of compounds that modulate cAMP/PKA signaling suggest a potential mechanism of action by which HMI-1a3 induces its PKA-mediated anticancer effects. These include, for example, adenylyl cyclase activation, through which forskolin has demonstrated anticancer potential via PKA activation (Follin-Arbelet et al., 2015). Another hypothetical mechanism of action is that HMI-1a3 could act as an inhibitor of phosphodiesterases, which control the amount of cAMP. Phosphodiesterase inhibitors specific for cAMP have been suggested to possess cancer drug–candidate potential (Peng et al., 2018). However, the contribution of other proteins containing a diacylglycerol-responsive C1 domain in the effects of HMI-1a3 cannot be excluded (Fig. 5).
Schematic representation of known and newly recognized targets of HMI-1a3 and the main findings of this study. MRCKα, myotonic dystrophy kinase–related CDC42-binding kinase-α.
When interpreting studies only utilizing cell lines, it is important to keep in mind that in two-dimensional models, the cell interactions do not accurately mimic the situation on the tissue level, in which several factors, such as tumor microenvironment, play a significant role (Peddareddigari et al., 2010; Fitzgerald et al., 2015). Thus, in further in vitro studies, primary cancer cells and three-dimensional spheroid models could better simulate clinical circumstances. An additional limitation of the current study is that the small-molecule compounds used as pharmacological tools may also target other kinases. Alternatively, PKA knockdown or overexpression could have been used to confirm the observed effects; however, compensatory expression changes and redundant roles of various kinases also complicate the interpretation of such studies.
In conclusion, our study identified PKA activation as a new anticancer mechanism of action for the PKC-targeted isophthalate derivative HMI-1a3 (Fig. 5). The mechanism by which HMI-1a3 activates PKA remains to be clarified in further studies. Furthermore, additional investigation is required to characterize the role of cAMP-PKA signaling in the other PKC-independent effects we have previously reported.
Acknowledgments
The authors thank Dr. Savithri Rangarajan and Dennis Buurman from PamGene International for scientific support and analysis of PamChip imaging data and Dr. Tuuli Karhu and Lotta Pohjolainen for advice and discussions. The Light Microscopy Unit (Institute of Biotechnology and Helsinki Institute of Life Science, University of Helsinki) is acknowledged for providing instrumentation and support for high-content analysis.
Authorship Contributions
Participated in research design: Tarvainen, Jäntti, Tuominen, Talman.
Conducted experiments: Tarvainen, Nunn.
Performed data analysis: Tarvainen, Nunn.
Wrote or contributed to the writing of the manuscript: Tarvainen, Jäntti, Tuominen, Talman.
Footnotes
- Received July 21, 2021.
- Accepted October 14, 2021.
The research was supported by the Magnus Ehrnrooth Foundation, Finnish Cultural Foundation, Orion Research Foundation sr, University of Helsinki Doctoral Programme in Drug Research, Jane and Aatos Erkko Foundation, Päivikki and Sakari Sohlberg Foundation, and Academy of Finland [Grant 321564].
No author has an actual or perceived conflict of interest with the contents of this article.
↵
 This article has supplemental material available at jpet.aspetjournals.org.
This article has supplemental material available at jpet.aspetjournals.org.
Abbreviations
- BrdU
- 5-bromo-2′-deoxyuridine
- CaMK 2
- Ca2+/calmodulin-dependent protein kinase type 2
- CDKL
- cyclin-dependent kinase-like
- CRC
- colorectal cancer
- DAPI
- 4′,6-diamidino-2-phenylindole
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- GSK-3
- glycogen synthase kinase 3
- HCA
- high-content analysis
- HMI-1a3
- 5-(hydroxymethyl)isophthalate 1a3
- JNK
- c-Jun N-terminal kinase
- LDH
- lactate dehydrogenase
- LNCaP
- lymph node carcinoma of the prostate
- MAPK
- mitogen-activated protein kinase
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- PARP
- poly(ADP-ribose) polymerase
- PKA
- protein kinase A
- PKC
- protein kinase C
- PMA
- phorbol 12-myristate 13-acetate
- PRAK
- p38-activated protein kinase
- RT
- room temperature
- STK
- Ser/Thr kinase
- VASP
- vasodilator-stimulated phosphoprotein
- Copyright © 2021 The Author(s)
This is an open access article distributed under the CC BY Attribution 4.0 International license.