Abstract
Abstract ID 127654
Poster Board 453
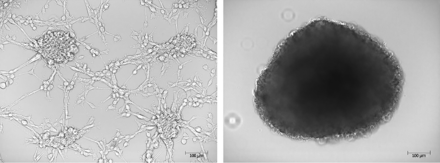
Introduction: Glioblastoma (GBM) is an aggressive brain cancer with high mortality. Small extracellular vesicles (sEVs) are nanoscale vesicles released by GBM to promote tumor survival. Development of GBM models to better understand the pro-tumor role of GBM sEVs is vital to improving patient outcomes. GBM mouse models have significant limitations that justify development of human 3D in vitro culture alternatives, but isolation of sEVs from traditional 3D gel matrix systems is limited. Alternatively, we developed a human U87 GBM 3D suspension spheroid culture system to generate scalable collection of easily retrievable GBM sEVs.
Hypothesis: The objective was to evaluate the biophysical and cancer pathway focused mRNA content differences between U87 sEVs produced by 2D adherent vs. 3D spheroid suspension culture.
Methods: sEVs were isolated from 2D adherent and 3D suspension cell culture using differential ultracentrifugation. Nanoparticle tracking analysis was used to determine the sEV diameter and zeta potential. RT-qPCR was used to determine fold regulation of 2D and 3D derived sEV mRNAs compared to their source cells and each other. Fold change significance was calculated using Student’s t-test of the replicate 2^(- Delta Ct) values for each gene in the control and treatment groups.
Results: 2D adherent and 3D spheroid suspension culture derived U87 sEVs do not differ significantly in terms of size at 249 +/- 153 and 265 +/- 316 nm respectively, or zeta potential (< -30 mV). A comparison of 3D vs. 2D (control) sEVs revealed enrichment of cancer immune-crosstalk mRNAs including CCL2, EGFR, STAT1 and TGF-β in 3D sEVs relevant to GBM survival. CCL2, STAT1 and EGFR facilitate tumor cell growth while TGF-β promotes tumor survival via immunosuppression.
Conclusions: The 3D U87 spheroid suspension model used in conjunction with 2D adherent culture might be used to streamline the identification of candidate GBM sEV-based biomarkers and therapeutic targets for GBM. This will be achieved by better simulating the 3D tumor mass microenvironment for sEV production and functional studies, and more accurately predicting functional mRNA and other sEV content. The 3D model can further be adapted for exploration of sEV crosstalk using co-culture systems, or multi-omics approaches.
Funding Support: UofL Cancer Education Program NIH/NCI (R25-CA134283-10), UofL faculty start-up funds to J. L. Hood, and UofL Hepatobiology and Toxicology COBRE NIH NIGMS P20GM113226.
Figure 1. Human U87 glioblastoma cells cultured in vitro. Bright-field microscopy of traditional 2D adherent cells (left) and 3D spheroid suspension cells (right) grown for 7 days. Scale bar = 100 μm
- Copyright © 2024 by The American Society for Pharmacology and Experimental Therapeutics