Abstract
Nonsense-mediated mRNA decay (NMD) is a mechanism, which selectively degrades transcripts carrying premature termination codons (PTCs) and a variety of physiologic transcripts containing NMD-inducing features. In a recent study, we have found variable NMD efficiency among nasal epithelial cells obtained from cystic fibrosis (CF) patients. This variability was found for CF transmembrane conductance regulator (CFTR) transcripts carrying the W1282X PTC, as well as for several NMD physiologic substrates. Here, we aimed to investigate the possibility that variability in NMD efficiency is a more generalized phenomenon and is not restricted to nasal epithelial cells. To investigate this possibility, we analyzed the NMD efficiency of both a CFTR constructs carrying the W1282X PTC and β-globin constructs carrying the NS39 PTC, in HeLa and MCF7 cells. Variability in NMD efficiency was found for both constructs between the cells, such that in HeLa cells the NMD was highly efficient and in MCF7 the efficiency was significantly lower. Moreover, similar differences in the efficiency of NMD were found for five endogenous NMD physiologic transcripts. Altogether, our results demonstrate existence of cells in which NMD of all transcripts is efficient, whereas others in which the NMD is less efficient, suggesting that the efficiency of NMD is an inherent character of cells. Our results also suggest that variability in the efficiency of NMD is a general phenomenon and is not restricted to nasal epithelial cells. As NMD affects the level of many transcripts, variability in the NMD efficiency might play a role as a genetic modifier of different cellular functions.
Similar content being viewed by others
Introduction
Nonsense-mediated mRNA decay (NMD) is a translation dependent post-transcriptional process in eukaryotic cells. This mechanism selectively degrades transcripts carrying premature translation-termination codons (PTCs), which are generally located >50–55 nucleotides upstream from downstream exon–exon junction. The purpose is to prevent the synthesis of non-functional or potentially deleterious proteins.1 Recently, it was found that NMD also degrades a variety of physiologic transcripts, which include transcripts with upstream open reading frame (uORF), transcripts containing introns in the 3′ untranslated region (UTR) and transcripts derived from alternative splicing.2, 3, 4, 5, 6
In a recent study, we have shown that NMD efficiency considerably varies among three cell lines derived from nasal epithelium of cystic fibrosis (CF) patients.7 We found variability in NMD efficiency for CF transmembrane conductance regulator (CFTR) transcripts carrying a disease-causing PTC, the W1282X mutation. Variability was also found for five physiologic NMD substrates. These included an alternatively spliced PTC-bearing transcript, RPL3; transcripts with introns in their 3′ UTR, SC35 1.6 kb and SC35 1.7 kb (two splicing isoforms); a transcript with a uORF, ASNS; and another bona fide NMD substrate with an unknown NMD-inducing feature, CARS. Our results demonstrated the existence of nasal epithelial cells in which NMD of all these analyzed transcripts was efficient and others in which the NMD was less efficient. Furthermore, downregulation of NMD increased the level of CFTR nonsense transcripts and led to enhanced CFTR chloride-channel activity in response to gentamicin.
Here we have investigated the possibility that variability in NMD efficiency is a more generalized phenomenon, and is not restricted to nasal epithelial cells. We demonstrate that the same PTC can elicit NMD with variable efficiencies when comparing two different epithelial cell lines, HeLa and MCF7. HeLa cells are derived from cervical carcinoma, whereas MCF7 are derived from breast adenocarcinoma. Variability in NMD efficiency was found for two different transcripts carrying disease-causing PTCs: CFTR constructs carrying the W1282X nonsense mutation and β-globin transcripts carrying the NS39 nonsense mutation. Variability was also found for five endogenous physiologic NMD substrates: RPL3; SC35 1.6 kb; SC35 1.7 kb; ASNS; and CARS. Altogether, our results suggest that the efficiency of NMD is an inherent character of cells. Furthermore, variability in the efficiency of NMD is a general phenomenon and is not restricted to nasal epithelial cells.
Materials and methods
Cell cultures
The human cell lines investigated in the study were of epithelial origin: HeLa, which was derived from cervical carcinoma and MCF7 from breast adenocarcinoma. The cell lines were grown in DMEM supplemented with 10% fetal calf serum.
Plasmid construction
Complete CFTR cDNA was inserted into pCMVbeta vector (ATCC), in which sequences of the beta-gal were deleted. We connected three PCR fragments generated from DNA extracted from a healthy individual carrying wild-type (WT) CFTR alleles. The PCR fragments contained exons 20 and part of its downstream intronic sequence (264 bp), exon 21 and part of its flanking introns (240 bp at intron 20 and 289 bp at intron 21) and exon 22 and part of its upstream intronic sequence (260 bp). The CFTR mutant construct was generated using the same cloning approach on DNA extracted from an individual homozygous for the W1282X mutation. Both constructs were sequenced and asides from the W1282X mutation no variation was identified in all exons and 199 bp of each intron.
β-Globin WT and mutant (NS39) constructs were described previously.8 An elongated human β-globin gene (wt300+e3) served as a control for transfection efficiency in all experiments, as described previously.9
Transfections
Transfections were performed by using an electroporation protocol (described in Agami and Bernards10) or by calcium-phosphate precipitation with standard methods. Similar transcript levels were obtained by using either of these transfection methods. The CFTR transfections were performed with either WT or mutant construct together with a plasmid encoding GFP (pEGFP, Clontech) for normalization. The β-globin transfections were performed with either WT or mutant construct, together with wt300+e3 construct for normalization. Transfections were performed using 1 or 2 μg of construct. The results showed no difference in transcript level with the different amounts of construct, excluding the possibility that transfection efficiency led to variability in transcript levels among the cell lines. Experiments were repeated at least three times.
RNA analysis
Total RNA was extracted using the RNeasy extraction kit (Qiagen) or Tri-Reagent-LS (MRC). RNA-less and reverse-transcriptase-less reactions were used as controls. For quantification of CFTR W1282X transcript levels in HeLa and MCF7 cells, which had been transfected with the CFTR constructs, we performed real-time PCR in the LightCycler (software version 3.5), using a FastStart DNA Master SYBR Green I kit (Roche Diagnostics). A GFP plasmid was used as a reference for transfection efficiency in each cell line. The level of mRNA transcribed from the WT or W1282X CFTR construct was normalized to the mRNA level of GFP. Subsequently, the ratio between the normalized mRNA level transcribed from the mutant and the WT constructs following cycloheximide (CHX) treatment was calculated and compared with this ratio in untreated cells. Sequence of the primer pairs are as follows: for pCFTR: forward (exon 20) ATGGTGTGTCTTGGGA, reverse (exon 22) ACAAGGACAAAGTCAAGC; for pEGFP: forward GCAACTACAAGACCCGC, reverse GTCGGCCATGATATAGACG. For each pair of primers, a standard curve was performed and annealing temperature was optimized in order to exclude PCR artifacts. Moreover, we included in the analysis only experiments in which the standard curves were as expected, to ensure accurate quantification.
For quantification of β-globin NS39 transcript levels in HeLa and MCF7 cells, transfected with the β-globin constructs, we performed Northern Blots with 1.5–5 μg of total cytoplasmic RNA according to standard protocols. Northern blots of total cytoplasmic RNA of transfected cells were probed with a β-globin-specific radiolabeled antisense cRNA. The control β-globin plasmid (wt300+e3) was used as a reference for transfection efficiency in each cell line. The level of mRNA transcribed from the WT or NS39 β-globin construct was normalized to the mRNA level of the β-globin wt300+e3 construct. Subsequently, the ratio between the normalized mRNA level transcribed from the mutant and the WT constructs following NMD inhibition was calculated and compared with this ratio in untreated cells.
The quantification of physiologic NMD substrates: RPL3; SC35 1.6 kb; SC35 1.7 kb; ASNS; and CARS, was performed as was previously described7 and as mentioned in the legend of Figure 3.
Effect of NMD inhibition on the transcript level of physiologic NMD substrates. The level of each analyzed transcript was measured by real-time PCR and normalized to that transcribed from GAPDH and the ratio between these normalized levels following treatment was calculated and compared with the ratio in untreated cells. (a–e) Effect of indirect NMD inhibition by CHX on the level of RPL3 (a), SC35 1.7 kb (b), SC35 1.6 kb (c), ASNS (d), and CARS (e) transcripts. (f–j) Effect of direct NMD inhibition by UPF1 downregulation on the level of RPL3 (f), SC35 1.7 kb (g), SC35 1.6 kb (h), ASNS (i), and CARS (j) transcripts. The increase in the level is shown as mean±SEM.
NMD inhibition
We indirectly inhibited NMD by treating the cells with 200 μg/ml CHX (Sigma) for 5 h. We directly inhibited NMD by using short-interfering RNA (siRNA) directed against UPF1. siRNA oligos for silencing UPF1 expression (Dharmacon) were those described by Mendell et al.11 siRNA oligo for luciferase (Dharmacon) was described by Gehring et al.12 Transfection conditions were described previously.12 Experiments were repeated at least three times.
Western blots
Total proteins (20–40 μg) were extracted from the different cell lines and were subjected to SDS-PAGE as was described previously.12 These proteins were probed with antibodies against UPF1 and tubulin.
Results
In order to investigate whether variability in NMD efficiency is a general phenomenon or restricted to nasal epithelial cells, we analyzed NMD efficiency in differently derived epithelial cells, HeLa and MCF7. We analyzed the efficiency of NMD for two different transcripts carrying disease-causing PTCs: CFTR constructs carrying the CF-causing PTC W1282X and the β-globin constructs carrying the β-thalassemia-causing PTC NS39. We also studied five endogenous physiologic NMD substrates: RPL3; SC35 1.6 kb; SC35 1.7 kb; ASNS; and CARS.
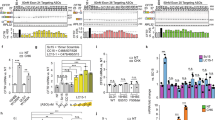
In our previous study, we analyzed the efficiency of NMD for endogenous CFTR transcripts carrying the W1282X nonsense mutation, in nasal epithelial cells derived from CF patients. As HeLa and MCF7 cells do not express CFTR gene, we transfected them with CFTR constructs carrying the W1282X PTC, which is located >55 nucleotides upstream to the final exon–exon junction, or the normal sequence (WT) (Figure 1a). We examined the levels of transfected W1282X transcripts following treatment with CHX, a general inhibitor of translation elongation and as NMD is a translation-dependent mechanism, CHX treatment leads indirectly to inhibition of NMD.13 A GFP plasmid was used as a reference for transfection efficiency in each cell line. The levels of mRNA transcribed from the WT or W1282X CFTR construct was normalized to the mRNA level of GFP. Subsequently, the ratio between the normalized mRNA levels transcribed from the mutant and the WT constructs following CHX treatment was calculated and compared with this ratio in untreated cells. The analysis showed that the fold increase differed between the cell lines such that in MCF7, a modest increase in the level was observed (1.3±0.03-fold), whereas in HeLa cells, the increase was ∼2.5 higher (3.2±0.27-fold) (Figure 1b). These results suggest that in MCF7 cells, the NMD of CFTR transcripts carrying the W1282X PTC is inefficient, as the transcript level showed only a marginal increase following the NMD indirect inhibition. The results further indicate that the NMD efficiency of CFTR might vary between different epithelial cell lines.
Effect of CHX treatment on the level of CFTR mRNA transcribed from constructs carrying the W1282X PTC. (a) A scheme of the WT (upper panel) and W1282X (lower panel) constructs, which contained all CFTR exons (marked in the boxes by numbers) and part of the intronic sequences between exons 20–22. The CMV promoter is marked by a thick horizontal arrow. (b) Real-time PCR analysis of CFTR transcripts following CHX treatment. The level of mRNA transcribed from CFTR constructs carrying either the normal sequence or the W1282X PTC was normalized to the mRNA level of GFP. The ratio between these normalized levels following CHX treatment was calculated and compared with the ratio in untreated cells. The fold increase in the level of CFTR W1282X transcripts is shown as mean±SEM.
We then analyzed the efficiency of NMD of β-globin constructs carrying the NS39 PTC or the normal sequence (WT) (Figure 2a), in HeLa and MCF7 cells. The levels of the NS39 transcripts were analyzed following transfection of the constructs and NMD inhibition. As a reference for transfection efficiency, we co-transfected the β-globin constructs with another β-globin construct, which included the normal sequence and a 300 bp exon (wt300+e3, control). The level of mRNA transcribed from β-globin constructs, carrying either the normal sequence or the NS39 PTC, was normalized to the mRNA level of the control construct. Subsequently, the ratio between the normalized mRNA level transcribed from the mutant and the WT constructs following NMD inhibition was calculated and compared with this ratio in untreated cells. Following CHX treatment, no increase in the level of NS39 transcripts was observed in MCF7 cells, whereas in HeLa cells a significant increase (4.4±0.20-fold) was observed (Figure 2b), similar to the variable effect of CHX on the level of CFTR W1282X transcripts in these cell lines. Subsequently, we inhibited NMD in these cells by siRNA duplexes against UPF1, a key factor that operates in the NMD machinery. As a control, we analyzed the effect of luciferase siRNA duplexes. Western blot analysis showed a sequence-specific and a very efficient downregulation of UPF1 expression in both HeLa and MCF7 cells (Figure 2c). The mRNA quantification results showed a similar effect to that observed following CHX treatment. Although in HeLa cells, a significant increase was observed (2.9±0.27-fold), in MCF7 cells, no increase was found (the difference between 43 and 31% or 0.7-fold increase following UPF1 downregulation is within the experimental variability) (Figure 2d). It is important to note that most of the knowledge regarding NMD mechanism comes from studies of the NS39 β-globin construct in HeLa cells. The increase in the level of NS39 β-globin transcripts found in our study in HeLa cells (∼3 to 4.5-fold) following UPF1 downregulation is similar to that found in previous studies.8, 14, 15 Together, these results show that the NMD efficiency of β-globin transcripts might vary between different cell lines, similar to the variability found for the CFTR in these cells. Specifically, both the CFTR and β-globin transcripts were more efficiently degraded in HeLa than in MCF7 cells.
Effect of NMD inhibition on the level of β-globin mRNA transcribed from constructs carrying the NS39 PTC. (a) A scheme of the WT (upper panel) and NS39 (lower panel) constructs, which contained all β-globin exons (marked in the boxes by numbers) and part of the intronic sequences. The CMV promoter is marked by a thick horizontal arrow. (b) Northern blot analysis of β-globin transcripts following CHX treatment. The level of mRNA transcribed from β-globin constructs carrying either the normal sequence or the NS39 PTC was normalized to the mRNA level of the control construct. The ratio between these normalized levels following treatment was calculated and compared with the ratio in untreated cells. (c) Western blot analysis in HeLa (left panel) and MCF7 (right panel) following sequence-specific downregulation of UPF1. (d) Northern blot analysis of β-globin transcripts following UPF1 downregulation. mRNA quantification was performed as described in (b). The fold increase in the level of β-globin NS39 is shown as mean±SEM.
We further extended our analysis in HeLa and MCF7 cells and studied the efficiency of NMD for physiologic NMD substrates following CHX treatment or downregulation of UPF1. The NMD physiologic substrates included RPL3; SC35 1.6 kb and SC35 1.7 kb; ASNS; and CARS. For all five analyzed transcripts, the results show an increase in the level following CHX treatment, which widely varied between the cells (Figure 3a–e, Figure 4). In MCF7 cells, the increase in the level of each analyzed transcripts was significantly lower than that found in HeLa cells. Similar results were found following UPF1 downregulation (Figure 3f–j, Figure 4 and Figure 4a with Figure 4b). It is interesting to note that the consistent results, which were obtained by NMD inhibition using CHX treatment or UPF1 siRNA in HeLa and MCF7 cells, emphasize that variable uptake or metabolism of CHX between these two cell lines is not likely the basis for variability in NMD efficiency. The 0.7-fold increase following UPF1 downregulation in MCF7 cells for RPL3 and SC35 1.7 kb transcripts is within the experimental variability, and indicates no increase in the level of these transcripts. It is important to note that the fold increase following CHX treatment was significantly higher than that observed following UPF1 downregulation (compare Figure 3a–e with Figure 3f–j and Figure 4a with Figure 4b). We conjecture that this effect probably resulted from the incomplete NMD inhibition following UPF1 downregulation. Altogether, the results from CHX treatment and from UPF1 downregulation show variability in NMD efficiency between HeLa and MCF7 cells. The results further show that the NMD in MCF7 is relatively less efficient than in HeLa cells.
Discussion
Our results provide evidence that the efficiency of NMD is an inherent character of cells and may vary among different cells. Previous studies showed variability in NMD efficiency only for transcripts carrying disease-causing PTCs among cells derived from different tissues. For example, a different increase was obtained in HeLa and T cells stably transfected with TCR-β construct carrying a PTC, following treatment with CHX.16 Variability in NMD efficiency was also found among chondrocyte, lymphoblasts and bone cells of a Schmid metaphyseal chondrodysplasia patient carrying a PTC in the collagen X gene.17 Recently, an additional study reported variability in NMD efficiency for transcripts carrying a deletion in the ESCO2 gene, which generates a PTC, among different tissues from two unrelated Roberts syndrome fetuses.18 Variability in NMD efficiency of CYH2 transcripts carrying a PTC among different yeast strains was also reported.19 The results presented in the current study together with the results from our recent work7 show, for the CFTR and the β-globin transcripts carrying a disease-causing PTC and for five physiologic NMD substrates, that there are cells, such as HeLa, CFP15a, and CFP22a, in which NMD of all transcripts was efficient and other cells, such as MCF7 and CFP15b, in which the NMD was less efficient. It is important to note that there are cells derived from the same tissue between these two cell groups, suggesting that the observed variable NMD efficiency between them does not reflect tissue-specific variability. Moreover, these findings strongly indicate that besides the position of the PTC relative to the exon–exon junction suggested previously in other studies,1 there are additional factors, which play a role in regulating the efficiency of NMD. Hence, variability in NMD efficiency probably results from interactions between elements in transcripts, which are subject to NMD and factors in the NMD machinery or regulation. Single-nucleotide polymorphisms in genes encoding the NMD factors and their regulators, in factors from the exon junction complex, or in other translation termination factors can lead to variable efficiency of NMD among cells. Different expression levels of one or more of these factors might also contribute to the variability in NMD efficiency.
It is important to note that variability in NMD efficiency might have a role as a genetic modifier of different cellular functions. Our previous study showed an important role for NMD efficiency in the response to readthrough treatment and demonstrated the effect of variable NMD efficiency on the response. Following treatment with 50–200 μg/ml gentamicin, concentration-dependent chloride efflux was detected in CFP15a cells, whereas in IB3-1 cells no significant activation was observed. Only following downregulation of UPF1 together with high gentamicin concentrations (100 and 200 μg/ml), an impressive CFTR activation was observed in IB3-1 cells, whereas in CFP15a cells downregulation of UPF1 or UPF2 in the presence of gentamicin at all concentrations, resulted in an improved chloride-channel activity compared with gentamicin alone. Variable efficiency of NMD can also affect disease severity in the cases of truncated proteins either by gain-of-function or dominant-negative effects. This may explain the phenotypic variability among patients of Duchenne muscular dystrophy (severe) and Becker muscular dystrophy (mild), carrying the same PTC in the dystrophin gene.20 Variable efficiency of NMD can also alter the pattern of inheritance of mendelian diseases. A well-known example is the β-thalassemia in which PTCs at the 5′ of the β-globin gene trigger NMD and thus lead to a recessive trait of β-thalassemia,21, 22 whereas PTCs at the 3′ does not generally trigger NMD and thus lead to dominant form of the disease.23, 24, 25
To summarize, our study provide evidence for variable NMD efficiency among cells from the same cell type, and even from the same tissue, as a general phenomenon and an inherent character of a given cell. Thus, NMD might have a role as a genetic modifier and hence the outcome of many studies in human genetic diseases has to be considered in this light.
References
Nagy E, Maquat LE : A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem Sci 1998; 23: 198–199.
Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC : Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat Genet 2004; 36: 1073–1078.
Lewis BP, Green RE, Brenner SE : Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc Natl Acad Sci USA 2003; 100: 189–192.
He F, Li X, Spatrick P, Casillo R, Dong S, Jacobson A : Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5′ to 3′ mRNA decay pathways in yeast. Mol Cell 2003; 12: 1439–1452.
Mitrovich QM, Anderson P : Unproductively spliced ribosomal protein mRNAs are natural targets of mRNA surveillance in C. elegans. Genes Dev 2000; 14: 2173–2184.
Morrison M, Harris KS, Roth MB : smg mutants affect the expression of alternatively spliced SR protein mRNAs in Caenorhabditis elegans. Proc Natl Acad Sci USA 1997; 94: 9782–9785.
Linde L, Boelz S, Nissim-Rafinia M et al: Nonsense-mediated mRNA decay affects nonsense transcript levels and governs response of cystic fibrosis patients to gentamicin. J Clin Invest 2007; 117: 683–692.
Thermann R, Neu-Yilik G, Deters A et al: Binary specification of nonsense codons by splicing and cytoplasmic translation. EMBO J 1998; 17: 3484–3494.
Neu-Yilik G, Gehring NH, Thermann R, Frede U, Hentze MW, Kulozik AE : Splicing and 3′ end formation in the definition of nonsense-mediated decay-competent human beta-globin mRNPs. EMBO J 2001; 20: 532–540.
Agami R, Bernards R : Distinct initiation and maintenance mechanisms cooperate to induce G1 cell cycle arrest in response to DNA damage. Cell 2000; 102: 55–66.
Mendell JT, ap Rhys CM, Dietz HC : Separable roles for rent1/hUpf1 in altered splicing and decay of nonsense transcripts. Science 2002; 298: 419–422.
Gehring NH, Neu-Yilik G, Schell T, Hentze MW, Kulozik AE : Y14 and hUpf3b form an NMD-activating complex. Mol Cell 2003; 11: 939–949.
Carter MS, Doskow J, Morris P et al: A regulatory mechanism that detects premature nonsense codons in T-cell receptor transcripts in vivo is reversed by protein synthesis inhibitors in vitro. J Biol Chem 1995; 270: 28995–29003.
Lykke-Andersen J, Shu MD, Steitz JA : Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell 2000; 103: 1121–1131.
Danckwardt S, Neu-Yilik G, Thermann R, Frede U, Hentze MW, Kulozik AE : Abnormally spliced beta-globin mRNAs: a single point mutation generates transcripts sensitive and insensitive to nonsense-mediated mRNA decay. Blood 2002; 99: 1811–1816.
Carter MS, Li S, Wilkinson MF : A splicing-dependent regulatory mechanism that detects translation signals. EMBO J 1996; 15: 5965–5975.
Bateman JF, Freddi S, Nattrass G, Savarirayan R : Tissue-specific RNA surveillance? Nonsense-mediated mRNA decay causes collagen X haploinsufficiency in Schmid metaphyseal chondrodysplasia cartilage. Hum Mol Genet 2003; 12: 217–225.
Resta N, Susca FC, Di Giacomo MC et al: A homozygous frameshift mutation in the ESCO2 gene: evidence of intertissue and interindividual variation in Nmd efficiency. J Cell Physiol 2006; 209: 67–73.
Kebaara B, Nazarenus T, Taylor R, Atkin AL : Genetic background affects relative nonsense mRNA accumulation in wild-type and upf mutant yeast strains. Curr Genet 2003; 43: 171–177.
Kerr TP, Sewry CA, Robb SA, Roberts RG : Long mutant dystrophins and variable phenotypes: evasion of nonsense-mediated decay? Hum Genet 2001; 109: 402–407.
Forget BG, Benz Jr EJ, Skoultchi A, Baglioni C, Housman D : Absence of messenger RNA for beta globin chain in beta(0) thalassaemia. Nature 1974; 247: 379–381.
Baserga SJ, Benz Jr EJ : Nonsense mutations in the human beta-globin gene affect mRNA metabolism. Proc Natl Acad Sci USA 1988; 85: 2056–2060.
Thein SL, Hesketh C, Taylor P et al: Molecular basis for dominantly inherited inclusion body beta-thalassemia. Proc Natl Acad Sci USA 1990; 87: 3924–3928.
Kazazian Jr HH, Dowling CE, Hurwitz RL, Coleman M, Stopeck A, Adams III JG : Dominant thalassemia-like phenotypes associated with mutations in exon 3 of the beta-globin gene. Blood 1992; 79: 3014–3018.
Hall GW, Thein S : Nonsense codon mutations in the terminal exon of the beta-globin gene are not associated with a reduction in beta-mRNA accumulation: a mechanism for the phenotype of dominant beta-thalassemia. Blood 1994; 83: 2031–2037.
Acknowledgements
We thank T Danieli, M Goldberg, and YS Oren for useful advice on the CFTR plasmid constructions. The work was partially supported by a grant from Yissum to Batsheva Kerem and a DFG Grant to Andreas E Kulozik.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Linde, L., Boelz, S., Neu-Yilik, G. et al. The efficiency of nonsense-mediated mRNA decay is an inherent character and varies among different cells. Eur J Hum Genet 15, 1156–1162 (2007). https://doi.org/10.1038/sj.ejhg.5201889
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.ejhg.5201889
Keywords
This article is cited by
-
FMRP-mediated spatial regulation of physiologic NMD targets in neuronal cells
Genome Biology (2024)
-
Opitz syndrome: improving clinical interpretation of intronic variants in MID1 gene
Pediatric Research (2023)
-
Deciphering the nonsense-mediated mRNA decay pathway to identify cancer cell vulnerabilities for effective cancer therapy
Journal of Experimental & Clinical Cancer Research (2021)
-
Functional analysis of two novel TBX5 variants present in individuals with Holt–Oram syndrome with different clinical manifestations
Molecular Genetics and Genomics (2021)
-
Antisense suppression of the nonsense mediated decay factor Upf3b as a potential treatment for diseases caused by nonsense mutations
Genome Biology (2018)