Abstract
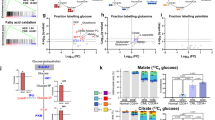
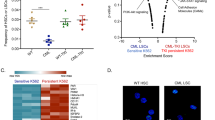
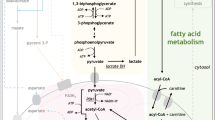
Treatment of chronic myeloid leukemia (CML) with imatinib mesylate and other second- and/or third-generation c-Abl-specific tyrosine kinase inhibitors (TKIs) has substantially extended patient survival1. However, TKIs primarily target differentiated cells and do not eliminate leukemic stem cells (LSCs)2,3,4. Therefore, targeting minimal residual disease to prevent acquired resistance and/or disease relapse requires identification of new LSC-selective target(s) that can be exploited therapeutically5,6. Considering that malignant transformation involves cellular metabolic changes, which may in turn render the transformed cells susceptible to specific assaults in a selective manner7, we searched for such vulnerabilities in CML LSCs. We performed metabolic analyses on both stem cell–enriched (CD34+ and CD34+CD38−) and differentiated (CD34−) cells derived from individuals with CML, and we compared the signature of these cells with that of their normal counterparts. Through combination of stable isotope–assisted metabolomics with functional assays, we demonstrate that primitive CML cells rely on upregulated oxidative metabolism for their survival. We also show that combination treatment with imatinib and tigecycline, an antibiotic that inhibits mitochondrial protein translation, selectively eradicates CML LSCs both in vitro and in a xenotransplantation model of human CML. Our findings provide a strong rationale for investigation of the use of TKIs in combination with tigecycline to treat patients with CML with minimal residual disease.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Druker, B.J. et al. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N. Engl. J. Med. 355, 2408–2417 (2006).
Graham, S.M. et al. Primitive, quiescent, Philadelphia-positive stem cells from patients with chronic myeloid leukemia are insensitive to STI571 in vitro. Blood 99, 319–325 (2002).
Corbin, A.S. et al. Human chronic myeloid leukemia stem cells are insensitive to imatinib despite inhibition of BCR-ABL activity. J. Clin. Invest. 121, 396–409 (2011).
Hamilton, A. et al. Chronic myeloid leukemia stem cells are not dependent on Bcr-Abl kinase activity for their survival. Blood 119, 1501–1510 (2012).
Holyoake, T.L. & Helgason, G.V. Do we need more drugs for chronic myeloid leukemia? Immunol. Rev. 263, 106–123 (2015).
Abraham, S.A. et al. Dual targeting of p53 and c-MYC selectively eliminates leukaemic stem cells. Nature 534, 341–346 (2016).
Tennant, D.A., Durán, R.V. & Gottlieb, E. Targeting metabolic transformation for cancer therapy. Nat. Rev. Cancer 10, 267–277 (2010).
Rowley, J.D. A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature 243, 290–293 (1973).
Groffen, J. et al. Philadelphia chromosomal breakpoints are clustered within a limited region, bcr, on chromosome 22. Cell 36, 93–99 (1984).
Konopka, J.B., Watanabe, S.M. & Witte, O.N. An alteration of the human c-abl protein in K562 leukemia cells unmasks associated tyrosine kinase activity. Cell 37, 1035–1042 (1984).
Rousselot, P. et al. Imatinib mesylate discontinuation in patients with chronic myelogenous leukemia in complete molecular remission for more than 2 years. Blood 109, 58–60 (2007).
Mahon, F.X. et al. Discontinuation of imatinib in patients with chronic myeloid leukaemia who have maintained complete molecular remission for at least 2 years: the prospective, multicentre Stop Imatinib (STIM) trial. Lancet Oncol. 11, 1029–1035 (2010).
Ross, D.M. et al. Safety and efficacy of imatinib cessation for CML patients with stable undetectable minimal residual disease: results from the TWISTER study. Blood 122, 515–522 (2013).
Simsek, T. et al. The distinct metabolic profile of hematopoietic stem cells reflects their location in a hypoxic niche. Cell Stem Cell 7, 380–390 (2010).
Takubo, K. et al. Regulation of glycolysis by Pdk functions as a metabolic checkpoint for cell cycle quiescence in hematopoietic stem cells. Cell Stem Cell 12, 49–61 (2013).
Yu, W.M. et al. Metabolic regulation by the mitochondrial phosphatase PTPMT1 is required for hematopoietic stem cell differentiation. Cell Stem Cell 12, 62–74 (2013).
Ito, K. et al. A PML–PPAR-δ pathway for fatty acid oxidation regulates hematopoietic stem cell maintenance. Nat. Med. 18, 1350–1358 (2012).
Carracedo, A., Cantley, L.C. & Pandolfi, P.P. Cancer metabolism: fatty acid oxidation in the limelight. Nat. Rev. Cancer 13, 227–232 (2013).
Sullivan, L.B. et al. Supporting aspartate biosynthesis is an essential function of respiration in proliferating cells. Cell 162, 552–563 (2015).
Birsoy, K. et al. An essential role of the mitochondrial electron transport chain in cell proliferation is to enable aspartate synthesis. Cell 162, 540–551 (2015).
Cardaci, S. et al. Pyruvate carboxylation enables growth of SDH-deficient cells by supporting aspartate biosynthesis. Nat. Cell Biol. 17, 1317–1326 (2015).
Norberg, E. et al. Differential contribution of the mitochondrial translation pathway to the survival of diffuse large B-cell lymphoma subsets. Cell Death Differ. 24, 251–262 (2017).
Skrtić, M. et al. Inhibition of mitochondrial translation as a therapeutic strategy for human acute myeloid leukemia. Cancer Cell 20, 674–688 (2011).
Mackay, G.M., Zheng, L., van den Broek, N.J. & Gottlieb, E. Analysis of cell metabolism using LC–MS and isotope tracers. Methods Enzymol. 561, 171–196 (2015).
Karvela, M. et al. ATG7 regulates energy metabolism, differentiation and survival of Philadelphia-chromosome-positive cells. Autophagy 12, 936–948 (2016).
Acknowledgements
The authors would like to dedicate this work to the memory of Prof. Tessa Holyoake who sadly passed away recently. Tessa was a dedicated clinician and an enthusiastic scientist who combined both aspects of her work for patients' benefit. She will be gravely missed.
We thank all patients and healthy donors who contributed samples and National Health Service (NHS) Greater Glasgow and Clyde Biorepository; A. Hair for sample processing; T. Gilbey and T. Harvey for cell sorting; N. Van Den Broek and G. MacKay for technical assistance; and A. King for editorial work. This study was supported by Cancer Research UK; the Cancer ResearchUK Glasgow Centre (C596/A18076) and the Biological Services Unit facilities at the Cancer Research UK Beatson Institute (C596/A17196); Medical Research Council/AstraZeneca project grants (MR/K014854/1); the Glasgow Experimental Cancer Medicine Centre (ECMC), which is funded by Cancer Research UK and by the Chief Scientist's Office (Scotland); the Howat Foundation and Friends of Paul O'Gorman; the Bloodwise Specialist Programme (14033); the Kay Kendall Leukaemia Fund (KKLF) (KKL501 and KKL698); Lady Tata International Award; and Leuka. G.V.H. is a KKLF Intermediate Research Fellow, Leadership Fellow and John Goldman Fellow.
Author information
Authors and Affiliations
Contributions
E.M.K., G.V.H. and E.G. wrote the manuscript. E.M.K., P.B., T.L.H., G.V.H. and E.G. designed experiments and interpreted data. E.M.K. and P.B. performed experiments and analyzed data. A.M.M. and K.D. assisted with the in vivo work. S.T. provided the custom-formulated culture medium. T.L.H. and G.V.H. provided primary cells. G.V.H. and E.G. supervised the project. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
E.G. is a founder and shareholder of MetaboMed, Ltd. T.L.H. has previously received research support from Bristol-Myers Squibb and Novartis.
Supplementary information
Supplementary Figures
Supplementary Figures 1–8. (PDF 39660 kb)
Supplementary Table 1
Comparative steady-state metabolomics analysis of patient-matched CD34+ and CD34− CML cells measured by LC–MS. (XLSX 18 kb)
Supplementary Table 2
Comparative steady-state metabolomics analysis of CD34+ CML and normal CD34+ cells measured by LC–MS. (XLSX 16 kb)
Rights and permissions
About this article
Cite this article
Kuntz, E., Baquero, P., Michie, A. et al. Targeting mitochondrial oxidative phosphorylation eradicates therapy-resistant chronic myeloid leukemia stem cells. Nat Med 23, 1234–1240 (2017). https://doi.org/10.1038/nm.4399
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.4399