Abstract
A fraction of ribosomes engaged in translation will fail to terminate when reaching a stop codon, yielding nascent proteins inappropriately extended on their C termini. Although such extended proteins can interfere with normal cellular processes, known mechanisms of translational surveillance1 are insufficient to protect cells from potential dominant consequences. Here, through a combination of transgenics and CRISPR–Cas9 gene editing in Caenorhabditis elegans, we demonstrate a consistent ability of cells to block accumulation of C-terminal-extended proteins that result from failure to terminate at stop codons. Sequences encoded by the 3′ untranslated region (UTR) were sufficient to lower protein levels. Measurements of mRNA levels and translation suggested a co- or post-translational mechanism of action for these sequences in C. elegans. Similar mechanisms evidently operate in human cells, in which we observed a comparable tendency for translated human 3′ UTR sequences to reduce mature protein expression in tissue culture assays, including 3′ UTR sequences from the hypomorphic ‘Constant Spring’ haemoglobin stop codon variant. We suggest that 3′ UTRs may encode peptide sequences that destabilize the attached protein, providing mitigation of unwelcome and varied translation errors.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Klauer, A. A. & van Hoof, A. Degradation of mRNAs that lack a stop codon: a decade of nonstop progress. Wiley Interdiscip. Rev. RNA 3, 649–660 (2012)
Hamby, S. E., Thomas, N. S., Cooper, D. N. & Chuzhanova, N. A meta-analysis of single base-pair substitutions in translational termination codons (‘nonstop’ mutations) that cause human inherited disease. Hum. Genomics 5, 241–264 (2011)
Williams, I., Richardson, J., Starkey, A. & Stansfield, I. Genome-wide prediction of stop codon readthrough during translation in the yeast Saccharomyces cerevisiae. Nucleic Acids Res. 32, 6605–6616 (2004)
Falini, B. et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N. Engl. J. Med. 352, 254–266 (2005)
Hollingsworth, T. J. & Gross, A. K. The severe autosomal dominant retinitis pigmentosa rhodopsin mutant Ter349Glu mislocalizes and induces rapid rod cell death. J. Biol. Chem. 288, 29047–29055 (2013)
Vidal, R. et al. A stop-codon mutation in the BRI gene associated with familial British dementia. Nature 399, 776–781 (1999)
Vidal, R. et al. A decamer duplication in the 3′ region of the BRI gene originates an amyloid peptide that is associated with dementia in a Danish kindred. Proc. Natl Acad. Sci. USA 97, 4920–4925 (2000)
Pang, S. et al. A novel nonstop mutation in the stop codon and a novel missense mutation in the type II 3beta-hydroxysteroid dehydrogenase (3beta-HSD) gene causing, respectively, nonclassic and classic 3β-HSD deficiency congenital adrenal hyperplasia. J. Clin. Endocrinol. Metab. 87, 2556–2563 (2002)
Clegg, J. B., Weatherall, D. J. & Milner, P. F. Haemoglobin Constant Spring—a chain termination mutant? Nature 234, 337–340 (1971)
Namy, O., Duchateau-Nguyen, G. & Rousset, J. P. Translational readthrough of the PDE2 stop codon modulates cAMP levels in Saccharomyces cerevisiae. Mol. Microbiol. 43, 641–652 (2002)
Inada, T. & Aiba, H. Translation of aberrant mRNAs lacking a termination codon or with a shortened 3′-UTR is repressed after initiation in yeast. EMBO J. 24, 1584–1595 (2005)
Shibata, N. et al. Degradation of stop codon read-through mutant proteins via the ubiquitin-proteasome system causes hereditary disorders. J. Biol. Chem. 290, 28428–28437 (2015)
Capone, J. P., Sharp, P. A. & RajBhandary, U. L. Amber, ochre and opal suppressor tRNA genes derived from a human serine tRNA gene. EMBO J. 4, 213–221 (1985)
Ahier, A. & Jarriault, S. Simultaneous expression of multiple proteins under a single promoter in Caenorhabditis elegans via a versatile 2A-based toolkit. Genetics 196, 605–613 (2014)
Doronina, V. A. et al. Site-specific release of nascent chains from ribosomes at a sense codon. Mol. Cell. Biol. 28, 4227–4239 (2008)
Jan, C. H., Friedman, R. C., Ruby, J. G. & Bartel, D. P. Formation, regulation and evolution of Caenorhabditis elegans 3’UTRs. Nature 469, 97–101 (2011)
Arribere, J. A. et al. Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics 198, 837–846 (2014)
Ingolia, N. T., Ghaemmaghami, S., Newman, J. R. S. & Weissman, J. S. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223 (2009)
Yen, H.-C. S., Xu, Q., Chou, D. M., Zhao, Z. & Elledge, S. J. Global protein stability profiling in mammalian cells. Science 322, 918–923 (2008)
Liebhaber, S. A. & Kan, Y. W. Differentiation of the mRNA transcripts originating from the alpha 1- and alpha 2-globin loci in normals and alpha-thalassemics. J. Clin. Invest. 68, 439–446 (1981)
Torabi, N. & Kruglyak, L. Genetic basis of hidden phenotypic variation revealed by increased translational readthrough in yeast. PLoS Genet. 8, e1002546 (2012)
Steneberg, P. & Samakovlis, C. A novel stop codon readthrough mechanism produces functional Headcase protein in Drosophila trachea. EMBO Rep. 2, 593–597 (2001)
Freitag, J., Ast, J. & Bölker, M. Cryptic peroxisomal targeting via alternative splicing and stop codon read-through in fungi. Nature 485, 522–525 (2012)
Eswarappa, S. M. et al. Programmed translational readthrough generates antiangiogenic VEGF-Ax. Cell 157, 1605–1618 (2014)
True, H. L. & Lindquist, S. L. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature 407, 477–483 (2000)
Waterston, R. H. A second informational suppressor, sup-7 X, in Caenorhabditis elegans. Genetics 97, 307–325 (1981)
Laski, F. A., Ganguly, S., Sharp, P. A., RajBhandary, U. L. & Rubin, G. M. Construction, stable transformation, and function of an amber suppressor tRNA gene in Drosophila melanogaster. Proc. Natl Acad. Sci. USA 86, 6696–6698 (1989)
Hudziak, R. M., Laski, F. A., RajBhandary, U. L., Sharp, P. A. & Capecchi, M. R. Establishment of mammalian cell lines containing multiple nonsense mutations and functional suppressor tRNA genes. Cell 31, 137–146 (1982)
Brenner, S. The genetics of Caenorhabditis elegans. Genetics 77, 71–94 (1974)
Okkema, P. G., Harrison, S. W., Plunger, V., Aryana, A. & Fire, A. Sequence requirements for myosin gene expression and regulation in Caenorhabditis elegans. Genetics 135, 385–404 (1993)
Granato, M., Schnabel, H. & Schnabel, R. pha-1, a selectable marker for gene transfer in C. elegans. Nucleic Acids Res. 22, 1762–1763 (1994)
Mello, C. C., Kramer, J. M., Stinchcomb, D. & Ambros, V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10, 3959–3970 (1991)
Stinchcomb, D. T., Shaw, J. E., Carr, S. H. & Hirsh, D. Extrachromosomal DNA transformation of Caenorhabditis elegans. Mol. Cell. Biol. 5, 3484–3496 (1985)
Mango, S. E., Lambie, E. J. & Kimble, J. The pha-4 gene is required to generate the pharyngeal primordium of Caenorhabditis elegans. Development 120, 3019–3031 (1994)
Zhong, M. et al. Genome-wide identification of binding sites defines distinct functions for Caenorhabditis elegans PHA-4/FOXA in development and environmental response. PLoS Genet. 6, e1000848 (2010)
Venolia, L. & Waterston, R. H. The unc-45 gene of Caenorhabditis elegans is an essential muscle-affecting gene with maternal expression. Genetics 126, 345–353 (1990)
Ao, W. & Pilgrim, D. Caenorhabditis elegans UNC-45 is a component of muscle thick filaments and colocalizes with myosin heavy chain B, but not myosin heavy chain A. J. Cell Biol. 148, 375–384 (2000)
Hodgkin, J., Papp, A., Pulak, R., Ambros, V. & Anderson, P. A new kind of informational suppression in the nematode Caenorhabditis elegans. Genetics 123, 301–313 (1989)
Hodgkin, J. A. & Brenner, S. Mutations causing transformation of sexual phenotype in the nematode Caenorhabditis elegans. Genetics 86, 275–287 (1977)
Mapes, J., Chen, J.-T., Yu, J.-S. & Xue, D. Somatic sex determination in Caenorhabditis elegans is modulated by SUP-26 repression of tra-2 translation. Proc. Natl Acad. Sci. USA 107, 18022–18027 (2010)
Anderson, P. & Brenner, S. A selection for myosin heavy chain mutants in the nematode Caenorhabditis elegans. Proc. Natl Acad. Sci. USA 81, 4470–4474 (1984)
Eide, D. & Anderson, P. The gene structures of spontaneous mutations affecting a Caenorhabditis elegans myosin heavy chain gene. Genetics 109, 67–79 (1985)
ENCODE Project Consortium. The ENCODE (ENCyclopedia Of DNA Elements) project. Science 306, 636–640 (2004)
Pédelacq, J.-D., Cabantous, S., Tran, T., Terwilliger, T. C. & Waldo, G. S. Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 24, 79–88 (2006)
Hopp, T. P. et al. A short polypeptide marker sequence useful for recombinant protein identification and purification. Nat. Biotechnol. 6, 1204–1210 (1988)
Field, J. et al. Purification of a RAS-responsive adenylyl cyclase complex from Saccharomyces cerevisiae by use of an epitope addition method. Mol. Cell. Biol. 8, 2159–2165 (1988)
Stadler, M. & Fire, A. Wobble base-pairing slows in vivo translation elongation in metazoans. RNA 17, 2063–2073 (2011)
Morlan, J. D., Qu, K. & Sinicropi, D. V. Selective depletion of rRNA enables whole transcriptome profiling of archival fixed tissue. PLoS One 7, e42882 (2012)
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013)
Nagalakshmi, U. et al. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320, 1344–1349 (2008)
Agarwal, V., Bell, G. W., Nam, J. W. & Bartel, D. P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 4, 1–38 (2015)
Miller, D. M. III, Ortiz, I., Berliner, G. C. & Epstein, H. F. Differential localization of two myosins within nematode thick filaments. Cell 34, 477–490 (1983)
Thompson, O. et al. The million mutation project: a new approach to genetics in Caenorhabditis elegans. Genome Res. 23, 1749–1762 (2013)
Kyte, J. & Doolittle, R. F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132 (1982)
Acknowledgements
We thank the Fire Laboratory for critical reading of the manuscript, C. Frøkjær-Jensen and K. Artiles for technical expertise, and T. Schedl, T. Inada, C. Joazeiro, L. Ling, A. Nager, and N. Spies for discussions. A. Sapiro and B. Li were instrumental in developing the RNA-seq2 protocol. This work was supported by grants from NIH R01GM37706, T32HG000044 (G.T.H.), 1DP2HD084069-01 (M.C.B.), 5F32GM112474-02 (J.A.A.), Walter and Idun Berry Foundation (E.S.C.), and NSF DGE-114747 (C.H.L.).
Author information
Authors and Affiliations
Contributions
J.A.A., E.S.C., and A.Z.F. designed C. elegans experiments. J.A.A. and E.S.C. conducted C. elegans experiments. N.J. developed the RNA-seq2 protocol. J.A.A. performed computational analyses. J.A.A. conducted experiments in human cell lines, as designed and aided by J.A.A., G.T.H., C.H.L., M.C.B., and A.Z.F. J.A.A. and A.Z.F. wrote the paper with help from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Sequencing data are available at Sequence Read Archive (SRP064516).
Reviewer Information Nature thanks J. S. Butler, M. Yarus and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Extended data figures and tables
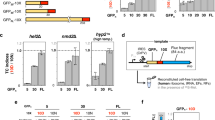
Extended Data Figure 1 Distribution of C-terminal extensions upon stop codon readthrough.
Annotations and genomes were as described in Supplementary Methods. Each 3′ UTR was translated starting one codon after the stop codon until the next in-frame stop codon. For metazoans, counting was performed in three different ways: including only genes for which exactly one 3′ UTR was annotated (blue), counting each annotated 3′ UTR separately (green), or counting each gene once and splitting gene counts with multiple 3′ UTRs equally amongst the 3′ UTR isoforms (red). ‘Nonstop’ denotes 3′ UTRs for which no stop codon was encountered before the poly(A) tail. For each species the distribution of next in-frame stop codons was calculated for 1,000 nucleotide shuffling of 3′ UTR sequences for genes with a single 3′ UTR annotated, and 95% confidence interval shown (yellow). A similar ‘randomized’ distribution was obtained upon shuffling 3′ UTR sequences and preserving dinucleotide frequency. The frequency of stops immediately after the annotated stop codon (amino acid length 0) is highlighted with a blue arrow in each species. The distribution of peptide lengths follows an exponential decay curve, where the slope is related to the probability of encountering a stop codon at each position. In the simplest model, the probability of encountering a stop codon is constant throughout the 3′ UTR, accounting for the roughly linear shape of each plot (previously noted2,3). Notable exceptions are a tendency towards second in-frame stops in E. coli (blue arrow), and a tendency towards peptides >60 amino acids in length in all species. In E. coli, the enrichment towards longer downstream peptides is at least partially explained by the operonic layout of genes.
Extended Data Figure 2 Example quantification of the GFP:mCherry fluorescence ratios of images.
Images were taken under a broad excitation and emission filter to allow for simultaneous capture of GFP and mCherry fluorescence. Intensities of each pixel in the red and green channels were extracted in python. Unfiltered pixel intensities are shown as black dots. Pixels were filtered, background subtracted, and linear regression performed (red dots and line, see Methods). For simplicity, the green–red intensities from 1,000 random pixels are shown. The GFP:mCherry fluorescence ratio was taken as the slope of the linear regression line. 10 × objective.
Extended Data Figure 3 Readthrough regions confer a loss of superfolder GFP fluorescence.
Each of the indicated TerByP regions were inserted downstream of superfolder (sf) GFP, upstream of the let-858 3′ UTR. TerByP is the region after the annotated stop codon, up to and including the first in-frame stop codon in the 3′ UTR. Quantification was performed as described in Extended Data Fig. 2.
Extended Data Figure 4 Explanation of ‘shuffle’ sequences.
Trinucleotide codons from each TerByP region are colour-coded by gene (top). Codons were extracted and randomly shuffled in python. A codon was iteratively selected until a stop codon was encountered, defining shuffle1. The process was repeated twice more to define shuffle2 and shuffle3. The resulting shuffle peptides are a combination of all three TerByP regions. Lengths and colour-coding of codons for shuffle1–3 accurately reflect the sequences they are derived from.
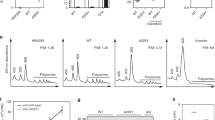
Extended Data Figure 5 RNA-seq and ribo-seq from unc-54 mutants.
a–c, RNA-seq (a) and ribosome footprint profiling (ribo-seq) (b) library mRNA counts, with summary counts (c) for the indicated strains and mRNAs. Libraries were prepared from L4 animals, as described Methods. ‘N2’ is wild type (PD1074, VC2010 (ref. 53)). unc-54(cc3389) bears a TAA (stop) to AAT (Asn) mutation, unc-54(TerByP). unc-54(e1301) has a GGA (Gly387) to AGA (Arg387) point mutation that confers a temperature-sensitive Unc phenotype with minimal discernible effects on UNC-54 protein levels. unc-54(e1301) was included as a control for the Unc phenotype of unc-54(cc3389), though e1301 confers a less severe Unc phenotype than cc3389. Values for unc-54 mRNA (blue) are highlighted throughout, and for comparison, three additional transcripts known to be at least partly expressed in the body-wall muscles are also highlighted: unc-87 (pink), unc-15 (green), and unc-22 (red).
Extended Data Figure 6 Ribo-seq of unc-54(cc3389) shows an unexceptional progression of ribosomes in the readthrough region.
a, Raw ribo-seq reads for unc-54(+) (blue) and unc-54(cc3389) (green) animals, plotted as read pile-ups. Mismatched bases are indicated with black bars. Location of the normal stop codon and the first in-frame stop codon are indicated with ‘TAA’ and dotted lines. The extension in unc-54(cc3389) is 30 amino acids. b, The number of ribo-seq reads in the last 30 codons, compared to the previous 30 codons, for all mRNAs. Linear regression was performed on all points (solid line), and twofold difference shown (dashed lines). c, The distribution of ribo-seq reads in the last 30 codons (90 nt) of unc-54(cc3389) is shown in green, and the 95% confidence interval (CI) for all open reading frames in dashed lines. d, The fraction of in-frame ribo-seq reads in the last 30 codons is plotted as a function of read counts in the last 30 codons, and unc-54(cc3389) highlighted. e, The distribution of read lengths in the last 30 codons of unc-54(cc3389), and all open reading frames (95% confidence interval, dashed lines). For b–d, reads were restricted to 28, 29, 30 nt lengths. For b–e, a 12 nt offset was performed for the ribosomal P-site, and read counts were derived solely from the unc-54(cc3389) ribo-seq library. For c and e, a minimum 15 read counts was imposed to obtain the 95% confidence interval from ‘all genes’.
Extended Data Figure 7 Lack of general conservation of coding potential downstream of stop codons in Caenorhabditis.
Whole-genome alignment of six nematode species with C. elegans genome assembly ce10/WS220 was obtained from the UCSC genome browser. For each annotated transcript, the aligned bases from the multiple species alignment were extracted and compared to the reference (C. elegans) genome. The left plot shows summary information of the alignment centred on annotated stop codons; the right plot shows the same centred on the first in-frame stop codon in 3′ UTRs. In red is the substitution frequency, that is, the number of mismatched bases divided by the number of aligned bases at a given position. The enrichment of ‘wobble’ position mutations is apparent as an increase in substitutions at the third position of each codon in the CDS. In green is the synonymous substitution frequency, that is, for codons beginning at a given position, the fraction of mutations that yield a synonymous substitution divided by all mutations at that position (synonymous and non-synonymous). The tendency to conserve amino acids in the CDS is apparent as a green spike at every in-frame codon. The change in substitution frequency and synonymous substitution frequency about the first in-frame stop codon (right plot) is due to a tendency for NTR codons to be conserved, and for AAN/AGN/GAN codons to not be conserved in 3′ UTRs, regardless of frame.
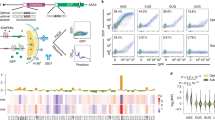
Extended Data Figure 8 Nucleotide and amino acid composition of readthrough regions (C. elegans).
CDSs and 3′ UTRs were analysed for various sequence properties. For simplicity, only genes and 3′ UTRs for which a single 3′ UTR was annotated were considered. Similar results were obtained with genes with multiple 3′ UTRs. a, Nucleotide frequency of CDS, 3′ UTR, and TerByP (region between annotated stop codon and first in-frame stop codon). b, Frequency of amino acids in all three possible frames for the TerByP region. 3′ UTRs were translated one codon past the stop codon of the CDS until the next in-frame stop codon, with nonstop 3′ UTRs ignored. Highlighted are codons with high G content (GGN, Gly) and high T content (TTY, Phe). c, TerByP regions tend to be hydrophobic, regardless of frame. Kyte–Doolittle score was used as a measure of hydrophobicity54. To reduce noise, only TerByP regions at least 10 amino acids long were considered. P value is from Kolmogorov–Smirnov test comparing CDSs and TerByP sequences (each frame has P value < 10e-293 for this comparison). As the TerByP sequences are shorter than CDSs on average, the distribution of TerByP hydrophobicity scores will tend to have higher variance than CDSs. Random portions of CDSs were taken, length-matched to TerByP frame zero peptide lengths. This was repeated 100 times, and the 95% confidence interval is shown (dashed lines, ‘CDS rands’). d, Hydrophobicity of the inserts is correlated with a negative effect on GFP fluorescence. The GFP:mCherry fluorescence ratio (Fig. 2b) was plotted against the maximum Kyte–Doolittle score in a six amino acid window for each insert. (Similar results were obtained using the Kyte–Doolittle score averaged across the entire sequence.) Mean (circle) and s.d. (bars) are shown. 3′-UTR-derived sequences are in blue, and non-3′-UTR-derived sequences are in red. To avoid redundancy or skewing of the data, in cases where multiple constructs were present with the same peptide sequence (for example, unc-54(TerByP), unc-54(TerByP, syn1), and unc-54(TerByP, syn2)), only the first of these was used. e, Hydrophobicity analysis of the TerByP extensions obtained by CRISPR–Cas9 engineering at the unc-22 and unc-54 loci. ‘+1/-1 TerByP’ denotes the gain or loss of a nucleotide, generating a late frameshift and allowing translation to proceed past the annotated stop codon out of frame with the upstream open reading frame. In each case, Kyte–Doolittle hydropathy was used to analyse the C-terminal appendage. The least phenotypically affected strain of the three is shown in bold.
Extended Data Figure 9 Nucleotide and amino acid composition of readthrough regions (H. sapiens).
a, b, Similar analysis of hydrophobicity as in Extended Data Fig. 8c, d, performed in humans.
Supplementary information
Supplementary Table 1
This table contains a list of strains used in the study. (XLSX 14 kb)
Supplementary Table 2
This table contains a list of plasmids used in the study. (XLSX 17 kb)
Supplementary Table 3
This table contains DNA oligos for rRNA digestion by RNaseH. (XLSX 7 kb)
Supplementary Information
This file contains supplementary text and gel source data for figure 3d. (PDF 704 kb)
Rights and permissions
About this article
Cite this article
Arribere, J., Cenik, E., Jain, N. et al. Translation readthrough mitigation. Nature 534, 719–723 (2016). https://doi.org/10.1038/nature18308
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature18308
This article is cited by
-
tRNA therapeutics for genetic diseases
Nature Reviews Drug Discovery (2024)
-
Ribosome biogenesis disruption mediated chromatin structure changes revealed by SRAtac, a customizable end to end analysis pipeline for ATAC-seq
BMC Genomics (2023)
-
Regulation of TRIB1 abundance in hepatocyte models in response to proteasome inhibition
Scientific Reports (2023)
-
Noncoding translation mitigation
Nature (2023)
-
Evolution of termination codons of proteins and the TAG-TGA paradox
Scientific Reports (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.