Abstract
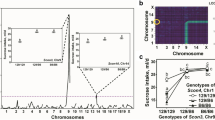
Short-term selective breeding starting from an F2 intercross of two inbred strains is a largely unexploited but potentially useful tool for quantitative trait locus (QTL) mapping. The selection lines can also serve as a valuable confirmation test of recornbinant inbred (RI) QTL results when the same two progenitor strains are used. Starting from an F2 from a C57BL/6J (B6) × DBA/2J (D2) cross (B6D2F2), this approach was used in a population of ~72 mice per generation bidirectionally selected for two-bottle choice 10% ethanol (alcohol) preference for four generations. The high-preference line diverged significantly from the low line in the first generation with a realized heritabittty of .32. By generation 4, the preference ratios in the high line were double those seen in the low line. Regions of the genome previously implicated by BXD RI QTL analysis as containing QTLs were searched using microsatellite markers. The test for the presence of QTLs was based on the divergence of marker allele frequencies in the two oppositely selected lines significantly exceeding that expected from random (genetic) drift and allele frequency estimation error. Combining the BXD and two-way selection line results, the most probable QTL was found on chromosome 3 (near the AdhI locus; LOD ~2.9), other probable QTLs were found with LOD 2.4–2.6.
Similar content being viewed by others
REFERENCES
Belknap, J. K., Machines, J. W., and McCleam, G. E. (1972). Ethanol-induced sleep time as a function of hepatic enzyme activities in mice. Physiol. Behav.9:453-457.
Belknap, J. K., Crabbe, J. C., and Young, E. R. (1993). Voluntary consumption of ethanol in 15 inbred mouse strains. Psychopharmacology112:503-510.
Belknap, J. K., Mogil, J. S., Helms, M. L., Richards, S. P., O'Toole, L. A., Bergeson, S. E., and Buck, K. J. (1995). Localization to chromosome 10 of a locus influencing morphine-induced analgesia in crosses derived from C57BL/'6 and DBA/2 mice. Life Sci. (Pharmacol. Lett.)57:PL117-PL124.
Belknap, J. K., Mitchell, S. R., O'Toole, L. A., Helms, M. L., and Crabbe, J. C. (1996). Type I and Type II error rates for quantitative trait loci (QTL) mapping studies using recombinant inbred mouse strains Behav. Genet.26:149-160.
Broadhurst, P. L. (1978). Drugs and the Inheritance of Behavior,Plenum, New York, pp. 131-149.
Crabbe, J. C., Phillips, T. J., Kosobud, A., and Belknap, J. K. (1990). Estimation of genetic correlation. Interpretation of experiments using selectively bred and inbred animals. Alcohol. Clin. Exp. Res.14:141-151.
Crabbe, J. C., Belknap, J. K., and Buck, K. J. (1994). Genetic animal models of alcohol and drug abuse. Science264: 1715-1723.
Darvasi, A,, and Soller, M. (1995). Advanced intercross lines, an experimental population for fine genetic mapping. Genetics141:199-1207.
DeFries, J. C., and Hegmann, J. P. (1970). Genetic analysis of open-field behavior. In Lindzey, G., and Thiessen, D. (eds.), Contributions to Behavior-Genetic Analysis: The Mouse as a Prototype, Appleton-Century-Crofts, New York, pp. 23-56.
Falconer, D. S. (1989). Introditction to Quantitative Genetics,Longman, New York.
Flint, J., Corley, R., DeFries, J. C., Fulker, D. W., Gray, J. A., Miller, S., and Collins, A. C. (1995). A simple genetic basis for a complex psychological trait in laboratory mice. Science 269:1432-1435.
Henderson, N. D. (1989). Interpreting studies that compare high-and low-selected lines on new characters. Behav. Genet. 19:473-502.
Hilbert, P., Lindpaintner, K., Beckmann, J. S., Serikawa, T., Soubrier, F., Dubay, C., Carrwright, P., De Gouyon, B., Julier, C., Takahasi, S., et al. (1991). Chromosomal mapping of two genetic loci associated with blood-pressure regulation in hereditary hypertensive rats. Nature353: 521-529.
Jacob, H. J., Lindpaintner, K., Lincoln, S. E., Kusumi, K., Bunker, R. K., Mao, Y.-P., Ganten, D., Dzau, V. J., and Lander, E. S. (1991). Genetic mapping of a gene causing hypertension in the stroke-prone spontaneously hypertensive rat. Cell67:213-224.
Johnson, T. E., DeFries, J. C., and Markel, P. (1992). Mapping quantitative trait loci for behavioral traits in the mouse. Behav. Genet.22:635-653.
Keightley, P. D., and Bulfleld, G. (1993). Detection of quantitative trait loci from frequency changes of marker alleles under selection. Genet. Res.62:195-203.
Lander, E. S., and Botstein, D. (1989). Mapping mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics121:185-199.
Lander, E. S., and Kruglyak, L. (1995). Genetic dissection of complex traits: Guidelines for interpreting and reporting linkage results. Nature Genet. 11:241-247.
Lander, E. S., and Schork, N. J. (1994). Genetic dissection of complex traits. Science265:2037-2048.
Lebowitz, R. J., Soller, M., and Beckmann, J. S. (1987). Traitbased analyses for the detection of linkage between marker loci and quantitative trait loci in crosses between inbred lines. Theor. Appl. Genet.73:556-562.
Manly, K. E., and Cudmore, R. (1995). Map Manager; A Programfor Genetic Mapping (v. 2.6.5), Roswell Park Cancer Institute, Buffalo, NY.
Phillips, T. J,, and Crabbe, J. C. (1991). Behavioral studies of genetic differences in alcohol action. In Crabbe, J. C., and Harris, R. A. (eds.), The Genetic Basis of Alcohol and Drug Actions, Plenum, New York, pp. 25-84.
Phillips, T. J., Crabbe, J. C., Metten, P., and Belknap, J. K. (1994). Localization of genes affecting alcohol drinking in mice. Alcohol. Clin. Exp. Res.18:931-941.
Rodgers, D. A. (1972). Factors underlying differences in alcohol preference in inbred strains of mice. In Kissin, B., and Begleiter, H. (eds,), The Biology of Alcoholism,Plenum, New York, pp. 107-130.
Rodgers, D. A., and McCleara, G, E. (1962), Alcohol preference in mice. In Bliss, E. L. (ed.), Roots of Behavior,Harper, New York, pp. 68-95.
Rodriguez, L. A., Plomin, R., Blizard, D. A., Jones, B. C., and McClearn, G. E. (1995). Alcohol acceptance, preference and sensitivity in mice. II. Quantitative trait loci mapping analysis using B X D recombinant inbred strains. Alcohol.Clin. Exp. Res. 19:367-373.
Schlesinger, K. (1966). Genetic and biochemical correlates of alcohol preference in mice. Am. J. Psychiat.122:767-773.
Shepard, J. R., Albersheim, P., and McClearn, G. E. (1968). Enzyme activities and ethanol preference in mice. Biochem. Genet.2:205-212.
Shepard, J. R., Albersheim, P., and McCleam, G. E. (1970). Aldehyde dehydrogenase and ethanol preference in mice. Biol. Chem.245:2875-2882.
Silver, L. M., Nadeau, J. H., and Goodfeliow, P. N. (1994). Encyclopedia of the mouse genome IV. Mammal. Genome5:S1-S295 (special issue).
Sokal, R. R., and Rohlf, F. J. (1981). Biometry,Freeman, San Francisco.
Taylor, B. A. (1978). Recombinant inbred strains: Use in gene mapping. In Morse, H. C. (ed.), Origins of Inbred Mice,Academic Press, New York, pp. 423-438.
Thomasson, H. R., Crabb, D. W., Edenberg, H. J., and Li, T.-K. (1993). Alcohol and aldehyde dehydrogenase polymorphisms and alcoholism. Behav. Genet.23:131-136.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Belknap, J.K., Richards, S.P., O'Toole, L.A. et al. Short-Term Selective Breeding as a Tool for QTL Mapping: Ethanol Preference Drinking in Mice. Behav Genet 27, 55–66 (1997). https://doi.org/10.1023/A:1025615409383
Issue Date:
DOI: https://doi.org/10.1023/A:1025615409383