Summary
In cancer chemotherapy neutropenia is a common dose-limiting toxicity. An ability to predict the neutropenic effects of cytotoxic agents based on proposed trial designs and models conditioned on previous studies would be valuable. The aim of this study was to evaluate the ability of a semi-mechanistic pharmacokinetic/pharmacodynamic (PK/PD) model for myelosuppression to predict the neutropenia observed in Phase I clinical studies, based on parameter estimates obtained from prior trials. Pharmacokinetic and neutropenia data from 5 clinical trials for diflomotecan and from 4 clinical trials for indisulam were used. Data were analyzed and simulations were performed using the population approach with NONMEM VI. Parameter sets were estimated under the following scenarios: (a) data from each trial independently, (b) pooled data from all clinical trials and (c) pooled data from trials performed before the tested trial. Model performance in each of the scenarios was evaluated by means of predictive (visual and numerical) checks. The semi-mechanistic PK/PD model for neutropenia showed adequate predictive ability for both anti-cancer agents. For diflomotecan, similar predictions were obtained for the three scenarios. For indisulam predictions were better when based on data from the specific study, however when the model parameters were conditioned on data from trials performed prior to a specific study, similar predictions of the drug related-neutropenia profiles and descriptors were obtained as when all data were used. This work provides further indication that modeling and simulation tools can be applied in the early stages of drug development to optimize future trials.
Similar content being viewed by others
Introduction
Cytotoxic chemotherapy suppresses the hematopoietic system affecting the host protective mechanisms. Among these hematological adverse effects, neutropenia is the most serious and usually the dose limiting toxicity [1]. Several pharmacokinetic/pharmacodynamic (PK/PD) models have been developed to describe the time-course of myelosuppression [2–6]. One of the more widely used is a semi-mechanistic PK/PD model developed by Friberg et al. 2002 [7]. This model describes the time course of myelosuppression after the administration of anticancer drugs by the use of system and drug related parameters. Consistency in its system-related parameters among a wide variety of anti-cancer agents and regimens has been demonstrated [7–16]. Recently, it has also been shown that this model could be used to predict the time-course of myelosuppression in humans, based on data obtained from experiments in rats [17].
In anti-cancer drug development, the ability to predict the incidence of neutropenia in subsequent trials, based on the trial design, and a model conditioned on one or more previously performed studies would be valuable. It has been shown for example, that a two-stage design of Phase I trials, in which data from an initial trial is used to condition the PK/PD model and to predict a safe starting dose for the subsequent trials may be applied safely, and is able to reduce the number of patients included at dose levels below the recommended dose [18]. As new data are accumulated during subsequent trials, model parameters can be updated to potentially provide less biased and more precise predictions of hematological toxicity.
Real data from the clinical development of two different anticancer drugs, diflomotecan and indisulam, were used in the current evaluation. These two drugs were given as anticancer monotherapy in their respective trials and therefore the neutropenia observed was a consequence of the administration of these drugs. Diflomotecan was the first homocamptothecin to enter clinical trials. Homocampotothecins are topoisomerase I inhibitors, that have greater ability to induce DNA cleavage mediated by topoisomerase I and a greater ability to stabilize topoisomerase I-DNA cleavable complexes when compared to the camptothecin-based topoisomerase I inhibitors [19, 20]. Indisulam is a cell cycle dependent anticancer drug that inhibits the cell-cycle at multiple checkpoints. The compound was shown to suppress the expression of cyclin E and phosphorylation of cyclin-dependent kinase 2 (CDK2), causing a disruption of the G1/S transition of the cell cycle [21]. Both compounds have shown neutropenia as the major dose limiting toxicity [22–24].
The aim of this study was to evaluate the ability of the model for hematological toxicity developed by Friberg et al. 2002 [7], to predict neutropenia early in the Phase I program of new anti-cancer agents, based on data gathered prior to the trial itself. The current investigation was performed assuming that Phase I development had occurred sequentially; i.e. trials investigating new regimens were only started once the current trial was completed. Model parameter estimates were obtained from: (a) data from each available trial independently, (b) pooled data from all clinical trials available or, (c) pooled data from the trials performed previously to the tested trial (Table 1). Using the parameter estimates from each scenario, absolute neutrophil count (ANC) profiles expected for each trial were simulated. The simulated profiles from each scenario were then compared with the observed data, thus providing a comparison of predictions made from: (a) model parameters derived only from the trial in question, (b) model parameters derived from all trials, and (c) model parameters derived only from those trials preceding the trial in question.
Methods
Study design and patients
Pharmacokinetic (PK) data and neutrophil counts from different clinical trials following administration of either diflomotecan or indisulam were available for the analysis. All participants provided written informed consent consistent with ICH-GCP (International conference on Harmonisation of Technical Requirements for Registration of Pharmaceuticals for Human Use—Good Clinical Practice) and local legislation, once the nature and the intention of the investigation were fully explained. The studies were performed in accordance with the Declaration of Helsinki and were approved by the institutional review board of the ethics committee at each study site. The characteristics of the patient population from the five diflomotecan clinical trials and the four indisulam clinical trials used for the current analysis are presented in Table 2.
Diflomotecan
Data from five Phase I clinical trials in advanced malignant tumors including a total of 111 patients were available. Patients were administered diflomotecan as intravenous infusion over 20 min on Day 1 (Study 1D) and on days 1, 2, 3, 4 and 5 (Study 4D) of a 21-day treatment cycle; and on Days 1, 7 and 14 of a 28-day treatment cycle (Study 2D). Patients in Studies 3D and 5D received an intravenous infusion on Day 1 followed by an oral administration given daily during Days 14 to 18. During the following cycles patients continued to receive an oral administration daily during five consecutive days every 21 days. The PK characteristics for intravenous and oral diflomotecan administrations have been described previously [10, 23].
Dose levels varied depending on the schedule and the type of administration (Table 2). Neutrophil counts were measured in peripheral blood every 3 to 7 days during all treatment cycles, measuring a total of 2,325 counts for the five studies.
Indisulam
Indisulam has been investigated in a Phase I clinical program, administered in four different treatment schedules [22, 25–27] from which PK and neutropenia data from a total of 150 patients were available. A population PK model has been developed previously to describe the disposition of indisulam in all four treatment schedules [28].
In Study 1I, patients were administered indisulam as a single 2 h infusion every 3 weeks, and in Study 2I indisulam was given in 5 daily 2-hour infusion, also every 3 weeks. In trial 3I, the regimen consisted of a four times weekly 1 h infusion, repeated every 6 weeks, while study 4I consisted of a continuous 120-hour infusion every 3 weeks. Neutrophil counts were recorded at study entry and at least once a week and, when feasible, twice a week, up to 2 weeks after the last infusion of indisulam. When hematological toxicity occurred, sampling was performed more frequently according to local guidelines. A total of 1,129 neutrophil counts were measured for the four studies.
Data analysis
The models were implemented in the non-linear mixed effects modeling software NONMEM version VI [29], in which the First Order Conditional Estimation (FOCE) method with the option INTERACTION was used for parameter estimation. For each drug the modeling was performed sequentially using the population modeling approach. First, the PK model was applied to obtain the individual parameter estimates for each study, and using these estimates the time course of ANC was modeled.
Pharmacokinetic model
The time course of the drug plasma concentrations for both anticancer agents was described previously by compartmental models parameterized in apparent volumes of distribution, and distribution and elimination clearances. The PK model for diflomotecan consisted of a three compartment model with a first order elimination [10]. Linear and dose-independent kinetics with an oral absolute bioavailability of 72 ± 59% of the total administered dose have been reported for this drug [23]. The indisulam PK model consisted of a three compartment model with saturable elimination from the central compartment [28]. Additional information on diflomotecan and indisulam PK can be found in previous publications [10, 23, 28].
Semi-mechanistic model for absolute neutrophil count
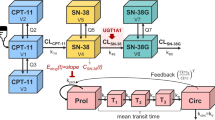
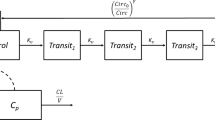
The semi-mechanistic model for chemotherapy-induced myelosuppression (Fig. 1) previously published by Friberg et al., 2002 [7] was fitted to the ANC vs. time data. This model consists of a system dependent and a drug dependent part. The system dependent part resembles in a simplified manner the underlying physiological processes determining the ANC in the circulation: (a) proliferation in a progenitor cell compartment, (b) maturation without proliferation, represented in the model by three transit compartments, (c) elimination from circulation, and (d) homeostatic regulation: i-iv can be described by the first order rate constant (kprol), the mean transit time (MTT), the first order rate constant of elimination from circulation (kcirc), and the feedback parameter (γ). In addition, Circ0 represents ANC at baseline. In the original model \( {k_{\Pr ol}} = {k_{circ}} = {k_{TR}} \), where kTR is the first order rate constant governing the transfer of immature cells between the transit compartments computed as \( \left( {n + 1} \right)/MTT \), n, being the number of transit compartments included in the model. The drug effect (EDRUG) was included in the model as \( {k_{\Pr ol}} \times \left( {1 - {E_{DRUG}}} \right) \) where EDRUG has the form of Slope × C (linear model), or \( {E_{MAX}} \times C/\left( {E{C_{50}} + C} \right) \) (EMAX model), where C represents the individual predicted plasma drug concentration, and Slope, EC50 and EMAX the PD model parameters to be estimated. In the cases where an EMAX model provided a better fit but EMAX and/or EC50 could not be estimated precisely, the following parameterization of the EMAX-model [30] was used to improve parameter precision:\( E{C_{50}} = {E_{MAX}}/SLP \), and \( {E_{DRUG}} = {E_{MAX}} \times C \times SLP/\left( {{E_{MAX}} + C \times SLP} \right) \), where EMAX and SLP are the parameters to be estimated.
Inter-patient variability (IPV) was modeled exponentially and residual variability was described using an additive random effect model. Neutrophil data were logarithmically transformed.
Model selection
The minimum value of the objective function provided by NONMEM, approximately equal to -2xlog likelihood (−2LL), was used as a guide during the model development. For two nested models a decrease in 3.84 points in -2LL for an extra parameter was considered significant at the 5% level.
Modeling strategy
The modeling strategy consisted of two steps: (a) parameter estimation from the different datasets (see below), and (b) evaluation of the predictive ability of the model.
-
(i)
Model parameter estimation
Independent analyses were performed for each drug and model parameter estimates were obtained for each of the following scenarios: Scenario A, data from each trial analyzed independently; Scenario B, pooled data from all available trials, and Scenario C, pooled data from the trials performed prior to the tested trial.
In Scenario A, five different datasets were studied for diflomotecan (Studies 1D, 2D, 3D, 4D and 5D were analyzed independently), and for indisulam four datasets were studied (Studies 1I, 2I, 3I and 4I were analyzed independently). Scenario B corresponded to one analysis of pooled data of all the studies for each drug. In Scenario C, for both diflomotecan and indisulam, it was assumed that the trials were performed sequentially, i.e. from 1D to 5D, and from 1I to 4I, thereby leading to four and three steps to be evaluated, respectively. This approach allowed having for each trial, from 2D to 5D, for diflomotecan and from 2I to 4I for indisulam, a set of parameters representing each of the scenarios be to be studied. A total of nine different parameter estimate sets for diflomotecan and seven for indisulam were obtained (Table 1).
-
(ii)
Evaluation of the prediction ability
The ability of the parameter estimates (obtained from each of the three different scenarios) to describe the observed data was tested by simulating five hundred datasets having the same doses, administration schedules and design characteristics as the original dataset and performing numerical and visual [31] predictive checks. Five hundred repetitions were used in the simulation exercise as the use of more repetitions did not relevantly alter the predicted outcome measures
Numerical predictive checks
The percentage of patients presenting neutropenia Grade 4 (%G4), and neutropenia Grade 3 or Grade 4 (%G3/4) were computed for each trial. Thereafter the overall mean and the 95% prediction intervals were calculated for each descriptor and compared to the same descriptors calculated from the raw data. This was then represented as a graphic, where %G4 or %G3/4 observed was plotted against the simulated %G4 or %G3/4 and its 95% prediction interval.
Visual predictive checks
The area covering the 95% confidence interval of the predicted median, the 5th and 95th percentiles of the prediction interval were plotted together with the median, and the 5th and 95th percentiles of the observed data. If the model described the data adequately the lines corresponding to the median, the 5th and 95th percentiles of the observed data would mainly fall in the area covering the 95% confidence interval.
For the analysis of simulated data, the R software (http://cran.r-project.org, version 2.6.0) was used. For the generation of visual predictive checks PsN (http://psn.sourceforge.net/) and Xpose version 4 (http://xpose.sourceforge.net/) were used.
Results
Diflomotecan
-
(i)
Model parameter estimation
For the nine datasets analyzed from the three explored scenarios the semi-mechanistic PK/PD model of neutropenia described the data adequately. Based on the minimum value of the objective function observations were considered best described by the reparameterized EMAX model (p < 0.01) except for the independent analysis of Studies 3D, 4D and 5D, where the drug effect was modeled using a linear function. The model parameter estimates are presented in Table 3. The data supported the estimation of IPV in Circ0, MTT and slope (for linear models) or SLP (for the reparameterized EMAX models). Inclusion of IPV in the feedback parameter was not statistically significant (p > 0.05) System- and drug-related parameters were consistent across the studies. Precision in parameter estimates increased when the number of pooled patients analyzed per study increased.
-
(ii)
Evaluation of the prediction ability
Numerical predictive checks
In Fig. 2 the mean values for the observed %G4 and %G3/4 neutropenia are shown against the mean %G4 and % G3/4 neutropenia calculated from the simulations. The observed value was within the prediction interval for the %G4 neutropenia for all evaluated situations, whereas for the %G3/4 neutropenia observations were outside the prediction interval in only one case i.e Study 4D in Scenario C (parameters estimates form previous studies were used in this case). For Study 4-D, in Scenario A and Scenario B the observations for %G3/4 were within the prediction intervals. However these two scenarios did not improve considerably the predictions when compared with Scenario C.
Relation between the percentage of Grade 4 or Grade ≥ 3 observed versus the percentage predicted for diflomotecan (top row) and indisulam (bottom row).Symbols represent the relation between the observed percentage of patients developing neutropenia (Grade 4 or Grade ≥ 3) and the median of the percentage of patients developing neutropenia (Grade 4 or Grade ≥ 3) from simulations. Lines represent the 95% interval of the predicted descriptor
Visual predictive checks
The visual predictive checks presented in Fig. 3 show that the description of data for each study was similar, independently of the set of model parameter estimates used for the simulations. For each study, the areas covering the 95% confidence interval of the predicted median, 5th and 95th percentiles were similar in each scenario.
Neutrophil count (logarithm) vs time profile after the administration of diflomotecan for the different studies. Median (solid line) and 95th and 5th percentiles (dashed line) of the observations. Dark gray areas cover the 90% CI of the median and light grey areas cover the 90%CI of the 95th and the 5th percentiles of the simulated profiles
Indisulam
-
(i)
Model parameter estimation
For the seven datasets analyzed from the three explored scenarios, the EMAX model did not provide significantly better fit, judged by the value of -2LL (p > 0.05) and visual predictive checks. Therefore, only linear models were implemented and estimated. As in previous analyses with this compound [32, 33], the Circ0 parameter was not estimated, instead observed baseline ANCs were used, thereby assuming no error in baseline measurement. Different methods of incorporating baseline with measurement error are available [34]. However when Circ0 was estimated, no relevant differences in predictions were observed and the VPCs for indisulam showed no apparent bias in the description of the baseline.
Data supported the estimation of IPV in MTT and slope in all estimations. The feedback parameter, γ could be estimated with reasonable precision for all scenarios (RSE <10%), and was between 0.116 and 0.190 (Table 4). The data did not support the estimation of IPV in the feedback parameter. The system- and drug-related parameters were consistent across the studies and the corresponding estimates are listed in Table 4.
-
(ii)
Evaluation of the prediction ability
Numerical predictive checks
In Fig. 2 (bottom row) the mean values for the observed %G4 and %G3/4 neutropenia are shown against the mean %G4 and % G3/4 neutropenia calculated from the simulations for indisulam. Predicted %G4 and %G3/4 were best correlated with the observed when the model was conditioned on data from the specific study only, as all prediction intervals contained the observed percentages. Predictions based on all data were comparable to those obtained using previously accumulated data. The observed value for the %G4 and the %G3/4 neutropenia was outside the prediction interval for the %G4 neutropenia for Study 4I and for the %G3/4 of the Study 3-I, when conditioned on all or previous data. For these scenarios, the semi-physiological model was not able to describe schedule-dependency adequately, and the occurrence of toxicity would have been significantly underestimated or overestimated.
Visual predictive checks
For all studies, model fit was best when the model was conditioned on data from the specific study alone (Fig. 4). Model fit was not equally good for the cases where the model was fitted on all data. However, when fitted on only previously accumulated data, the model fit was similar to when all data were used. For Study 4I, studying the continuous infusion, model misspecification was present when the model was conditioned on all or previous data: the observed lower boundary of the 90% prediction interval was significantly lower than predicted in several consecutive bins. When conditioned on data from the study itself, the misspecification was less apparent.
Neutrophil count (logarithm) vs time profile after the administration of indisulam for the different studies. Median (solid line), 95th and 5th percentiles (dashed line) of the observations. Dark gray areas cover the 95% CI of the median and light grey areas cover the 95% CI of the 95th and the 5th percentile of the simulated profiles
Discussion
The predictive ability of the PK/PD neutropenia model was evaluated for the anticancer drugs diflomotecan and indisulam. For both drugs simulations showed, visually and numerically, that a very similar description of data is achieved when data from all studies was used to condition the model, compared to the situation when data from the prior studies only was used. In most cases, such model performance also corresponded with the description of data provided by the model conditioned on the specific data of the study. Although each study had a different dose administration schedule, and a wide range of doses were studied throughout all the trials, the approach described here allowed for predictions based on model parameters obtained from prior trials. However an interesting fact seen for both of the drugs was that for Scenario C (use of previous data) the predictions did not turn out to be better as data were gained. For example the indisulam predictions for Study 4I based on previous data (i.e. Study 1I, 2I and 3I) were not better than predictions for Study 2I based only on Study 1I. This could be due to a schedule dependency in the development of toxicity that is not be captured by the model as Study 4I has a very different schedule (120 h infusion every 21 days) from the previous schedules tested. For diflomotecan a similar case was presented, where percentage of Grade 3/4 neutropenia for Studies 3D and 4D was not as well predicted as for Study 2I in Scenario C.
The semi-mechanistic model was successfully applied in all cases for both drugs, and in general precise parameters were provided. For diflomotecan in most of the cases an EMAX model fitted data better than a linear model, except for the low dose studies where it was not possible to fit an EMAX model. This was most likely due to the fact that these concentrations would correspond to the linear part of the response-concentration curve. In these linear models the difference for the slope parameter of 0.625 to 1.05 mL/ng (Studies 3D-5D) might be caused by the use of a reduced model due to the limited range of concentrations in those particular studies. However, when the EMAX model was selected the range in the SLP and EMAX parameters was smaller: 0.82–1.35 mL/ng and 5.33–6.46, respectively. In the manuscript by Trocóniz et al. [10], data obtained after administration of higher doses of diflomotecan were analyzed. Based on the results provided by the current evaluation where the EMAX model was identified as a better model than the linear one, the slope estimated in that case (0.144 mL/ng) should correspond to the upper (less steep) part of the effect vs concentration relationship.
Although system-related parameters obtained between studies and the two compounds were consistent, MTT estimates were in general higher for indisulam than for diflomotecan. A higher MTT estimate for indisulam in comparison with other drugs has been reported previously [9].
The critical aspect in the current approach resides in the extrapolation capability of model parameter estimates obtained from a specific study to previously unstudied schedules. Therefore, it was of importance to study the model performance when using the model from data not including the current data of the study (Scenario C). In general, the visual predictive checks showed that ANC time profiles obtained for that case were similar to the ones for the cases where the current study data were taken into account (Scenarios A and B), indicating that parameter estimates could be extrapolated. However, and in particular for Study 4I, the description of the ANC time profile was considerably better for the case in which only data from the study was used to condition the model (Scenario A). The administration schedule of Study 4I consisted of a 5 day long continuous infusion; all the other studies analyzed had an infusion duration of 2 h or less. For this study, besides the large difference in administration schedule, also differences in the studied patient population may have inflated the differences in predictions. For instance, Study 4I had the lowest number of patients included, the highest percentage female patients, the lowest median weight, and the lowest median baseline ANCs of all four studies. If only data from a low number of patients is available, a critical assessment of predictive ability is difficult, since of those patients, only a low number of patients will develop grade 3/4 toxicities. Therefore, calculation of confidence intervals of observed ANC counts for such a study is sensitive to the inclusion of a few outlying patients.
However, for both drugs, using data of all studies (Scenario B) provided very similar predictions as using only data from prior studies (Scenario C). For indisulam, predicting neutropenia toxicity based on data from previous analyses had shown to be only moderately accurate. Prediction could however only be improved when data from the specific study were used.
The predictive performance of the three sets of parameters was also compared for their ability to predict clinically relevant descriptors, i.e. the incidence of Grade 3 or Grade 4 neutropenia. Therefore, the percentage of patients developing either Grade 4 neutropenia, or Grade 3 neutropenia or higher, was calculated for both the observed data and the simulated data.
It could be expected that the percentage of neutropenia for a specific study would be better predicted by the model conditioned on the data from that specific study. This means that the left panels in Fig. 2 would present the best relationship between observed and predicted data. For some studies however, very similar predictions were seen regardless the set of model parameters used in the simulation (very similar predictions in all panels).
Predictions were more accurate for diflomotecan than for indisulam, especially for the % G4 neutropenia. A possible explanation for this result may be that in the diflomotecan trials a higher percentage of patients experienced neutropenia than in indisulam trials (Fig. 2), resulting in a better characterized dose response relationship.
Previous studies have already shown that estimated system-related model parameters are consistent across anti-cancer agents [7–16]. Furthermore, the predictive ability of this model has been also explored before in (i) a development of a tool that would be able to predict hematological toxicity in subsequent clinical trials, based on the study design and a model conditioned on one or more previously performed trials [18], and also in (ii) a method for interspecies scaling where time course of myelosuppression was predicted based on rat data [17], showing in both cases promising results. Further studies will have to verify if the same predictive ability applies to dose escalation studies.
Assuming that the Phase I development programs of studied drugs were performed sequentially, the current analysis explored the ability of the currently most used semi-mechanistic model of neutropenia to predict hematological effects in subsequent stages of the program, based on data gathered at earlier stages. The results presented in the current report are encouraging given the fact that for two drugs with different mechanism of action, and under very different dosing schedules, trials performed prior to a specific study gave a similar insight to the drug related-neutropenia as when using information from all studies retrospectively. It might be interesting to expand this research to other scenarios, such as the use of the PRIOR subroutine in NONMEM to introduce previous information, or the use of published system parameters in combination with drug related parameters estimated from previous studies.
This investigation provides further indication that modeling and simulation tools can be applied in the early stage of the drug development process to optimize future trials. Nevertheless, further studies should be done to determine the causes of the model-misspecification that led to the reduced predictive ability for some administration schedules.
References
Lyman GH, Dale DC, Crawford J (2003) Incidence and predictors of low dose-intensity in adjuvant breast cancer chemotherapy: a nationwide study of community practices. J Clin Oncol 21:4524–4531. doi:10.1200/JCO.2003.05.002
Minami H, Sasaki Y, Saijo N, Ohtsu T, Fujii H, Igarashi T, Itoh K (1998) Indirect-response model for the time course of leukopenia with anticancer drugs. Clin Pharmacol Ther 64:511–521. doi:10.1016/S0009-9236(98)90134-5
Zamboni WC, D’Argenio DZ, Stewart CF, MacVittie T, Delauter BJ, Farese AM, Potter DM, Kubat NM, Tubergen D, Egorin MJ (2001) Pharmacodynamic model of topotecan-induced time course of neutropenia. Clin Cancer Res 7:2301–2308
Panetta JC, Kirstein MN, Gajjar AJ, Nair G, Fouladi M, Stewart CF (2003) A mechanistic mathematical model of temozolomide myelosuppression in children with high-grade gliomas. Math Biosci 186:29–41
Vainstein V, Ginosar Y, Shoham M, Ranmar DO, Ianovski A, Agur Z (2005) The complex effect of granulocyte colony-stimulating factor on human granulopoiesis analyzed by a new physiologically-based mathematical model. J Theor Biol 234:311–327. doi:10.1016/j.jtbi.2004.11.026
Bulitta JB, Zhao P, Arnold RD, Kessler DR, Daifuku R, Pratt J, Luciano G, Hanauske AR, Gelderblom H, Awada A, Jusko WJ (2009) Multiple-pool cell lifespan models for neutropenia to assess the population pharmacodynamics of unbound paclitaxel from two formulations in cancer patients. Cancer Chemother Pharmacol 63:1035–1048. doi:10.1007/s00280-008-0828-1
Friberg LE, Henningsson A, Maas H, Nguyen L, Karlsson MO (2002) Model of chemotherapy-induced myelosuppression with parameter consistency across drugs. J Clin Oncol 20:4713–4721
Leger F, Loos WJ, Bugat R, Mathijssen RH, Goffinet M, Verweij J, Sparreboom A, Chatelut E (2004) Mechanism-based models for topotecan-induced neutropenia. Clin Pharmacol Ther 76:567–578. doi:10.1016/j.clpt.2004.08.008
van Kesteren C, Zandvliet AS, Karlsson MO, Mathot RA, Punt CJ, Armand JP, Raymond E, Huitema AD, Dittrich C, Dumez H, Roche HH, Droz JP, Ravic M, Yule SM, Wanders J, Beijnen JH, Fumoleau P, Schellens JH (2005) Semi-physiological model describing the hematological toxicity of the anti-cancer agent indisulam. Invest New Drugs 23:225–234. doi:10.1007/s10637-005-6730-3
Troconiz IF, Garrido MJ, Segura C, Cendros JM, Principe P, Peraire C, Obach R (2006) Phase I dose-finding study and a pharmacokinetic/pharmacodynamic analysis of the neutropenic response of intravenous diflomotecan in patients with advanced malignant tumours. Cancer Chemother Pharmacol 57:727–735. doi:10.1007/s00280-005-0112-6 [doi]
Latz JE, Karlsson MO, Rusthoven JJ, Ghosh A, Johnson RD (2006) A semimechanistic-physiologic population pharmacokinetic/pharmacodynamic model for neutropenia following pemetrexed therapy. Cancer Chemother Pharmacol 57:412–426. doi:10.1007/s00280-005-0077-5 [doi]
Puisset F, Alexandre J, Treluyer JM, Raoul V, Roche H, Goldwasser F, Chatelut E (2007) Clinical pharmacodynamic factors in docetaxel toxicity. Br J Cancer 97:290–296. doi:10.1038/sj.bjc.6603872
Kathman SJ, Williams DH, Hodge JP, Dar M (2007) A Bayesian population PK-PD model of ispinesib-induced myelosuppression. Clin Pharmacol Ther 81:88–94. doi:10.1038/sj.clpt.6100021
Brain EG, Rezai K, Lokiec F, Gutierrez M, Urien S (2008) Population pharmacokinetics and exploratory pharmacodynamics of ifosfamide according to continuous or short infusion schedules: an n = 1 randomized study. Br J Clin Pharmacol 65:607–610. doi:10.1111/j.1365-2125.2007.03095.x
Fetterly GJ, Grasela TH, Sherman JW, Dul JL, Grahn A, Lecomte D, Fiedler-Kelly J, Damjanov N, Fishman M, Kane MP, Rubin EH, Tan AR (2008) Pharmacokinetic/pharmacodynamic modeling and simulation of neutropenia during phase I development of liposome-entrapped paclitaxel. Clin Cancer Res 14:5856–5863. doi:10.1158/1078-0432.CCR-08-1046
Hansson EK, Wallin JE, Lindman H, Sandstrom M, Karlsson MO, Friberg LE (2009) Limited inter-occasion variability in relation to inter-individual variability in chemotherapy-induced myelosuppression. Cancer Chemother Pharmacol. doi:10.1007/s00280-009-1089-3
Friberg LE, Sandstrom M, Karlsson MO (2009) Scaling the time-course of myelosuppression from rats to patients with a semi-physiological model. Invest New Drugs. doi:10.1007/s10637-009-9308-7
Zandvliet AS, Karlsson MO, Schellens JH, Copalu W, Beijnen JH, Huitema AD (2009) Two-stage model-based clinical trial design to optimize phase I development of novel anticancer agents. Invest New Drugs. doi:10.1007/s10637-008-9216-2
Graham JS, Falk S, Samuel LM, Cendros JM, Evans TR (2009) A multi-centre dose-escalation and pharmacokinetic study of diflomotecan in patients with advanced malignancy. Cancer Chemother Pharmacol 63:945–952. doi:10.1007/s00280-008-0795-6
Lesueur-Ginot L, Demarquay D, Kiss R, Kasprzyk PG, Dassonneville L, Bailly C, Camara J, Lavergne O, Bigg DC (1999) Homocamptothecin, an E-ring modified camptothecin with enhanced lactone stability, retains topoisomerase I-targeted activity and antitumor properties. Cancer Res 59:2939–2943
Owa T, Yoshino H, Okauchi T, Yoshimatsu K, Ozawa Y, Sugi NH, Nagasu T, Koyanagi N, Kitoh K (1999) Discovery of novel antitumor sulfonamides targeting G1 phase of the cell cycle. J Med Chem 42:3789–3799
Raymond E, ten Bokkel Huinink WW, Taieb J, Beijnen JH, Faivre S, Wanders J, Ravic M, Fumoleau P, Armand JP, Schellens JH, European Organization for the Research and Treatment of Cancer Early Clinical Study Group (2002) Phase I and pharmacokinetic study of E7070, a novel chloroindolyl sulfonamide cell-cycle inhibitor, administered as a one-hour infusion every three weeks in patients with advanced cancer. J Clin Oncol 20:3508–3521
Gelderblom H, Salazar R, Verweij J, Pentheroudakis G, de Jonge MJ, Devlin M, van Hooije C, Seguy F, Obach R, Prunonosa J, Principe P, Twelves C (2003) Phase I pharmacological and bioavailability study of oral diflomotecan (BN80915), a novel E-ring-modified camptothecin analogue in adults with solid tumors. Clin Cancer Res 9:4101–4107
Scott L, Soepenberg O, Verweij J, de Jonge MJ, Th Planting AS, McGovern D, Principe P, Obach R, Twelves C (2007) A multicentre phase I and pharmacokinetic study of BN80915 (diflomotecan) administered daily as a 20-min intravenous infusion for 5 days every 3 weeks to patients with advanced solid tumours. Ann Oncol 18:569–575. doi:10.1093/annonc/mdl439
Punt CJ, Fumoleau P, van de Walle B, Faber MN, Ravic M, Campone M (2001) Phase I and pharmacokinetic study of E7070, a novel sulfonamide, given at a daily times five schedule in patients with solid tumors. A study by the EORTC-early clinical studies group (ECSG). Ann Oncol 12:1289–1293
Terret C, Zanetta S, Roche H, Schellens JH, Faber MN, Wanders J, Ravic M, Droz JP, EORTC Early Clinical Study Group (2003) Phase I clinical and pharmacokinetic study of E7070, a novel sulfonamide given as a 5-day continuous infusion repeated every 3 weeks in patients with solid tumours. A study by the EORTC Early Clinical Study Group (ECSG). Eur J Cancer 39:1097–1104
Dittrich C, Dumez H, Calvert H, Hanauske A, Faber M, Wanders J, Yule M, Ravic M, Fumoleau P (2003) Phase I and pharmacokinetic study of E7070, a chloroindolyl-sulfonamide anticancer agent, administered on a weekly schedule to patients with solid tumors. Clin Cancer Res 9:5195–5204
Van Kesteren C, Mathot RA, Raymond E, Armand JP, Dittrich C, Dumez H, Roche H, Droz JP, Punt C, Ravic M, Wanders J, Beijnen JH, Fumoleau P, Schellens JH, Early Clinical Studies Group of the European Organization for Research and Treatment of Cancer (2002) Population pharmacokinetics of the novel anticancer agent E7070 during four phase I studies: model building and validation. J Clin Oncol 20:4065–4073
Beal SL, Sheiner LB (1989–2006) NONMEM users guides. Icon development solutions, Ellicot City, Maryland, USA.
Schoemaker RC, van Gerven JM, Cohen AF (1998) Estimating potency for the Emax-model without attaining maximal effects. J Pharmacokinet Biopharm 26:581–593
Karlsson M, Holford NH (2008) A tutorial on visual predictive checks. Available: www.page-meeting.org/?abstract=1434.
Zandvliet AS, Schellens JH, Dittrich C, Wanders J, Beijnen JH, Huitema AD (2008) Population pharmacokinetic and pharmacodynamic analysis to support treatment optimization of combination chemotherapy with indisulam and carboplatin. Br J Clin Pharmacol 66:485–497. doi:10.1111/j.1365-2125.2008.03230.x
Zandvliet AS, Siegel-Lakhai WS, Beijnen JH, Copalu W, Etienne-Grimaldi MC, Milano G, Schellens JH, Huitema AD (2008) PK/PD model of indisulam and capecitabine: interaction causes excessive myelosuppression. Clin Pharmacol Ther 83:829–839. doi:10.1038/sj.clpt.6100344
Dansirikul C, Silber HE, Karlsson MO (2008) Approaches to handling pharmacodynamic baseline responses. J Pharmacokinet Pharmacodyn 35:269–283. doi:10.1007/s10928-008-9088-2
Acknowledgments
This research was supported with grants from the Swedish Cancer Society. Elena Soto received a PhD studentship from Boehringer Ingelheim Pharma GmbH & Co and Ron Keizer received a grant from the René Vogels Foundation / Dutch Society for Oncology.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Soto, E., Keizer, R.J., Trocóniz, I.F. et al. Predictive ability of a semi-mechanistic model for neutropenia in the development of novel anti-cancer agents: two case studies. Invest New Drugs 29, 984–995 (2011). https://doi.org/10.1007/s10637-010-9437-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10637-010-9437-z